L10 DNA Polymerases
E.coli DNA Polymerase I
Discovered by A. Kornberg 1957.
$\alpha$ DNA-dependent DNA polymerase
5’ to 3’ polymerase activity
inherent 3′– 5´ and 5′– 3′ exonuclease activities
Application:
- Nick translation for labeling
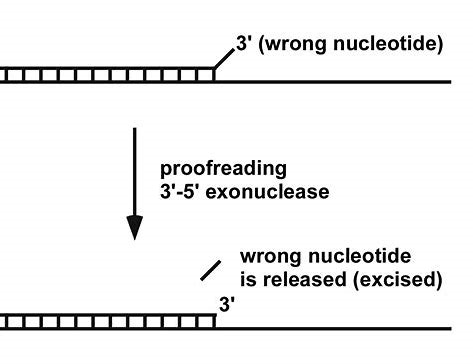
3’ to 5′ exonuclease activities – proofreading activity
5′ to 3’ exonuclease activities – chew up RNA primer during DNA replication.
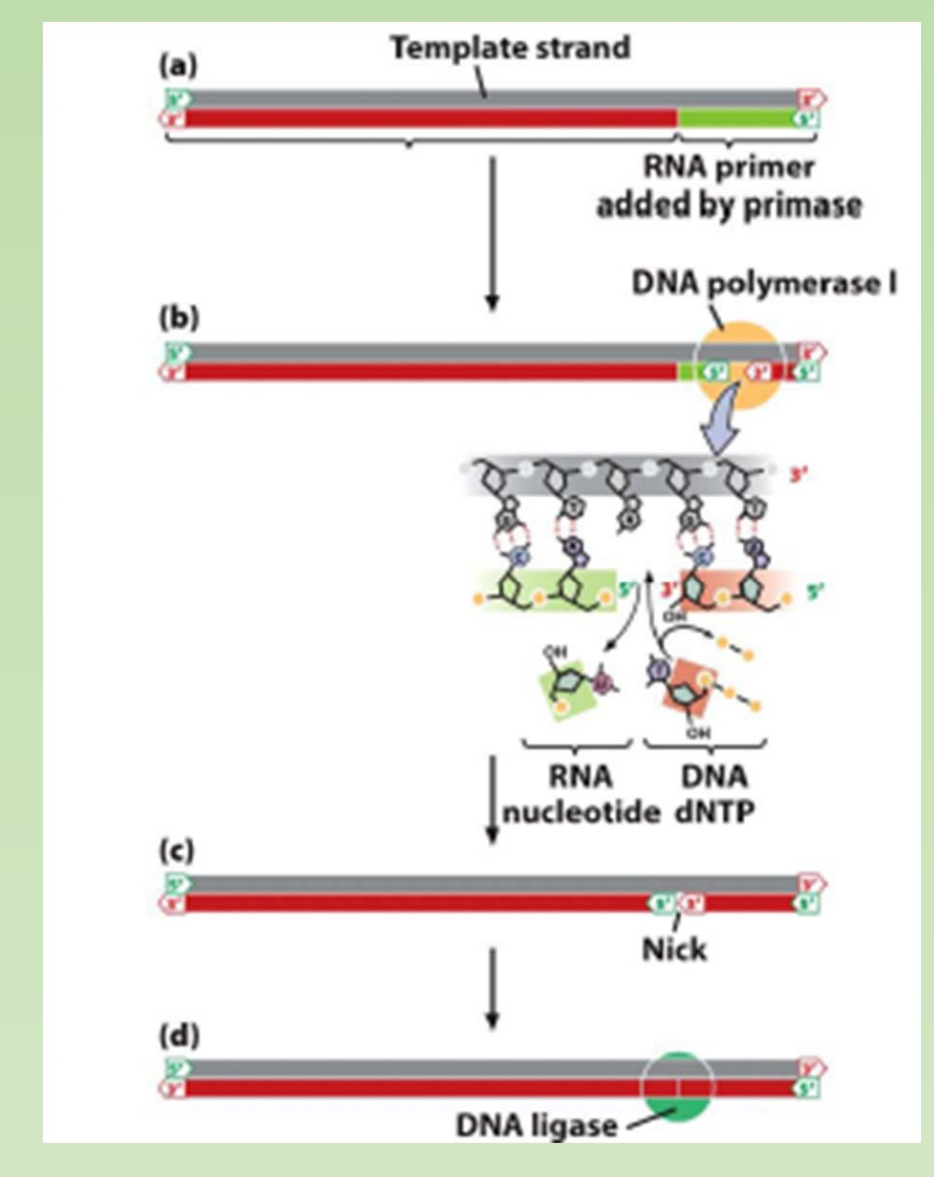
With DNA replication there is a leading strand which follows the replication fork and a lagging strand which goes in the other direction. Before DNA replication can begin Primase must lay down RNA primers so that DNA polymerase III can elongate the new strand by laying down new bases. Once elongation is complete there is the issue with having RNA primers are the start of the leading strand and also in between the Okazaki fragments on the lagging strand. This is where DNA polymerase I comes in. DNA polymerase I has a 5’-3’ exonuclease that chews up the RNA primer so that it can replace the RNA with DNA. Once it has done this ligase comes in to seal the nick
Klenow Fragment
DNA Polymerase I, Large (Klenow) Fragment
Without 5′– 3′ exonuclease activity
5’ to 3’ polymerase activity
3′ to 5´ exonuclease activity
Applications:
- Generates probes using random primers
- Second strand cDNA synthesis
- Dideoxy sequencing (less frequently used)
T4 DNA Polymerase
5’ to 3’ polymerase activity
Strong 3′ to 5’ exonuclease activity (much higher than DNA Pol I)
Without 5′– 3′ exonuclease activity
Applications:
- Gap filling, 5’ overhang filling
- 3’ overhang removal
- Probe labeling using replacement synthesis
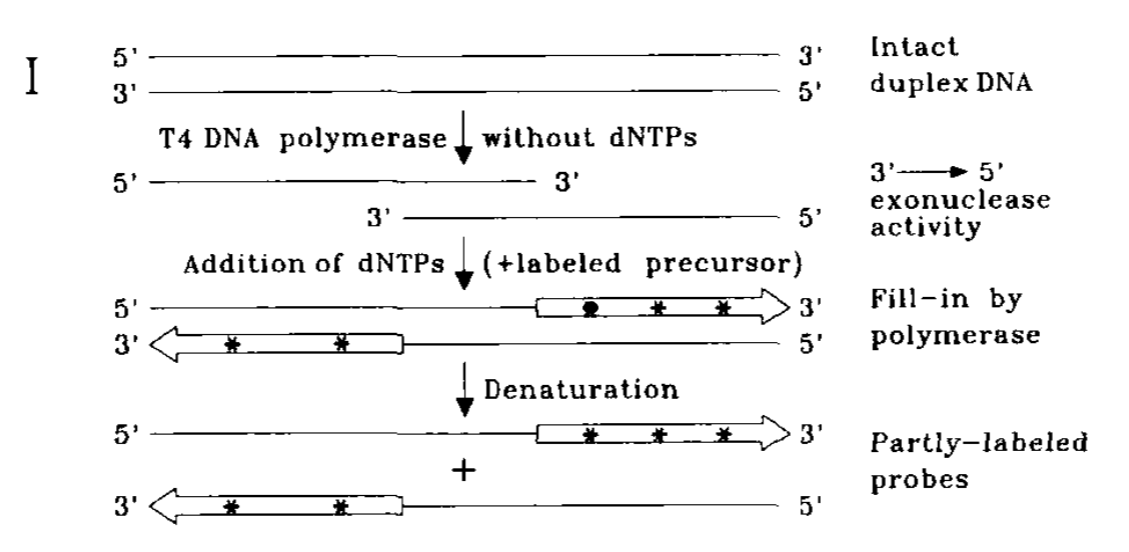
T4 DNA Pol behave like an 3’ to 5’ exonuclease in the absence of dNTPs
T7 DNA Polymerase
catalyze the DNA replication of T7 phage genome
High 5’ to 3’ polymerase activity (rapid extension rate)
High 3′ to 5´ exonuclease activity
Applications:
- Gap filling, 5’ overhang filling
- Synthesize long stretch of DNA (several kb)
- Native T7 DNA Pol is unsuitable for DNA sequencing, why?
- Engineered T7 DNA Pol are used in Sanger sequencing
phi29 DNA Polymerase
Replicative polymerase from the Bacillus subtilis phage phi29
5’ to 3’ polymerase activity (extremely processive, up to 70kb)
3′ to 5´ exonuclease activity
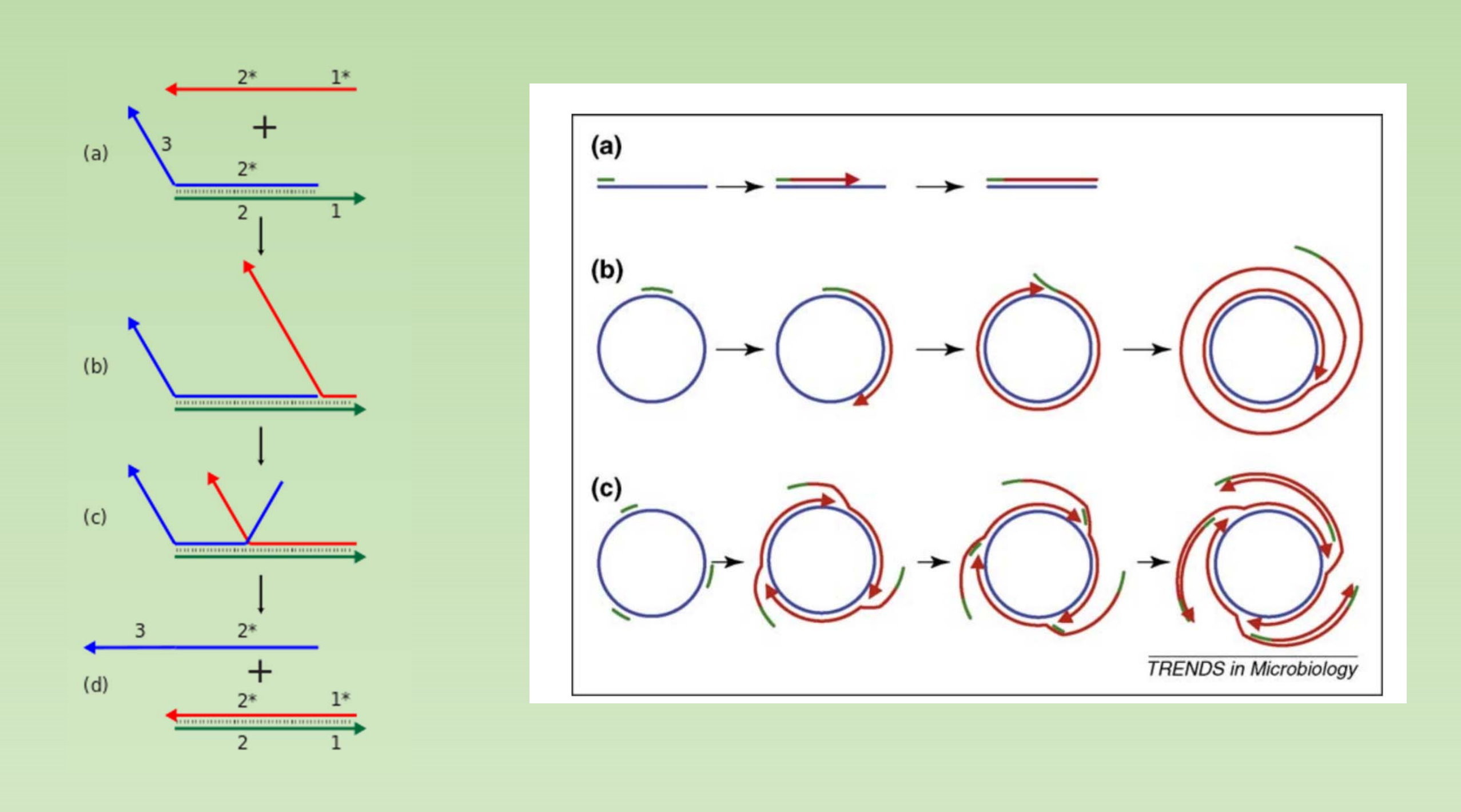
very high strand displacement activity (the ability to displace downstream DNA encountered during synthesis)
Applications:
- Ideal for rolling cycle amplification
- PacBio sequencing (smart-seq).
Reverse transcriptase
RNA directed DNA polymerase
AMV RT (Avian Myelobastosis Virus) & M-MLV RT (Moloney Murine Leukemia Virus), what’s the difference ?
AMV RT
- relatively insensitive to RNA secondary structure,
- Intrinsic RNase H activity, which degrades RNA of RNA/DNA hybrid and can cleave the RNA template if the RT pauses during synthesis.
- RNA template no longer available
- Difficult to obtain fragment longer than 5kb.
M-MLV RT
- Moloney Murine Leukemia Virus Reverse Transcriptase
- RNase H activity less than AMV-RT
- Different versions of RNase H– mutant available, idea for making long cDNA.
- Some RNase H- variants can work at 55 degree, can reduce the effect of RNA secondary structure
- Reaction is more sensitive to impurities of RNA samples.
- inhibited 50% by 5% (v/v) formamide, 17% (v/v) DMSO, 34% (v/v) glycerol, 15mM guanidine isothiocyanate, 1mM EDTA, 0.0025% SDS and 4µg/ml heparin
A comparison between different DNA polymerase

DNA polymerases
- E.coli DNA Pol I (Nick translation, 5’ to 3’ exonuclease activity)
- Klenow fragement (Random probe labeling)
- T4 DNA polymerase (High 3’ to 5’ exonuclease activity, Gibson Assembly
- T7 DNA polymerase (High polymerase activity, Engineered for DNA sequencing
- phi29 DNA polymerase (PacBio sequencing, Rolling Cycler Amplification
- Reverse transcriptase (AMV versus M-MLV, Rnase H- version).