L14 The application of Genetic Engineering
一、Genome Editing – The principle of CRISPR system
Genome editing pre-CRISPR: The development
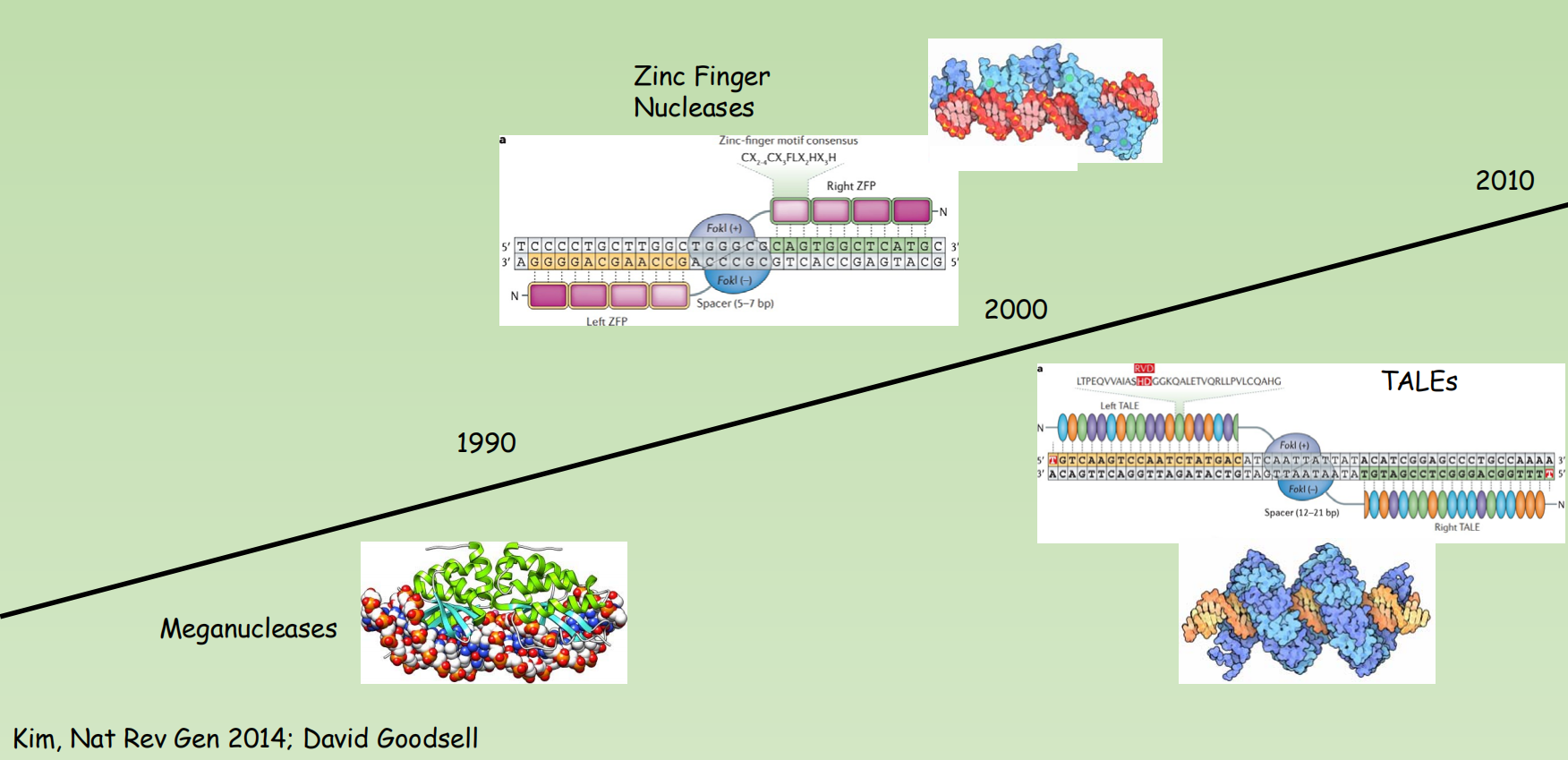
General principles
Editing the genome using
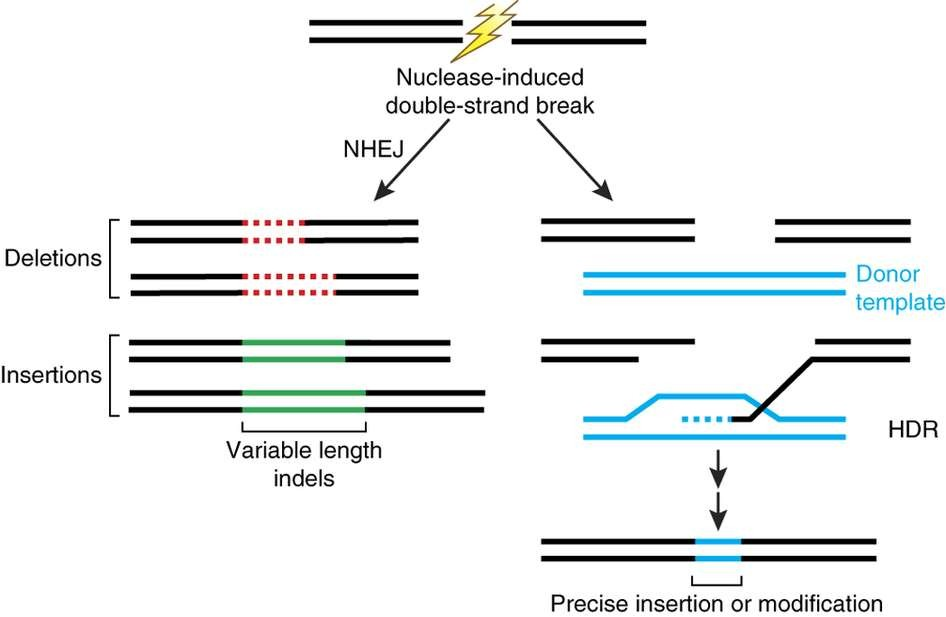
- Homology directed Repair(HDR) 同源定向修复
- Non Homologous End Joining (NHEJ) 非同源端链接
- Early approaches(早期方法): modifying genetic sequences using only homologous recombination.
NHEJ & HDR
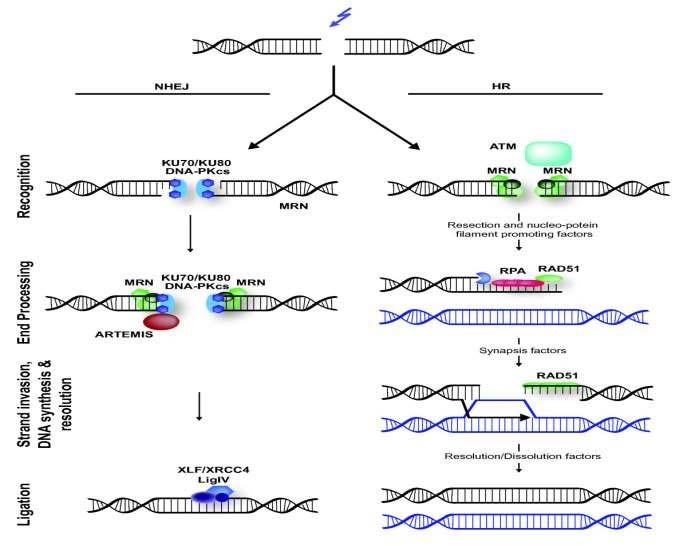
1. Homology directed repair (HDR)
Mechanism in cells to repair double strand DNA lesions
- The most common form homologous recombination
- Only used by the cell when there is a homologue piece of DNA present in the nucleus
Mostly in G2 and late S phase - Important for suppressing cancer
maintains genomic stability by repairing broken DNA strands
- Assumed to be error free because of the use of a template
2. Non-homologous end joining (NHEJ)
Repairs double-strand breaks in DNA in the absence of homologous templates
- Occurs in G0/G1 and early S-phases
non-homologous: the break ends directly ligated
non-homologous end joining: Moore and Haber(1996)
utilizes short homologous DNA sequences called microhomologies to guide repair
- Imprecise repair when the overhangs are not compatible
Lead to translocations and telomere fusion: hallmarks of tumor cells
CRISPRs
Clustered Regularly Interspaced Short Palindromic Repeats
Genetic elements that bacteria use as a kind of acquired immunity to protect against viruses
1. How CRISPRs Works ?
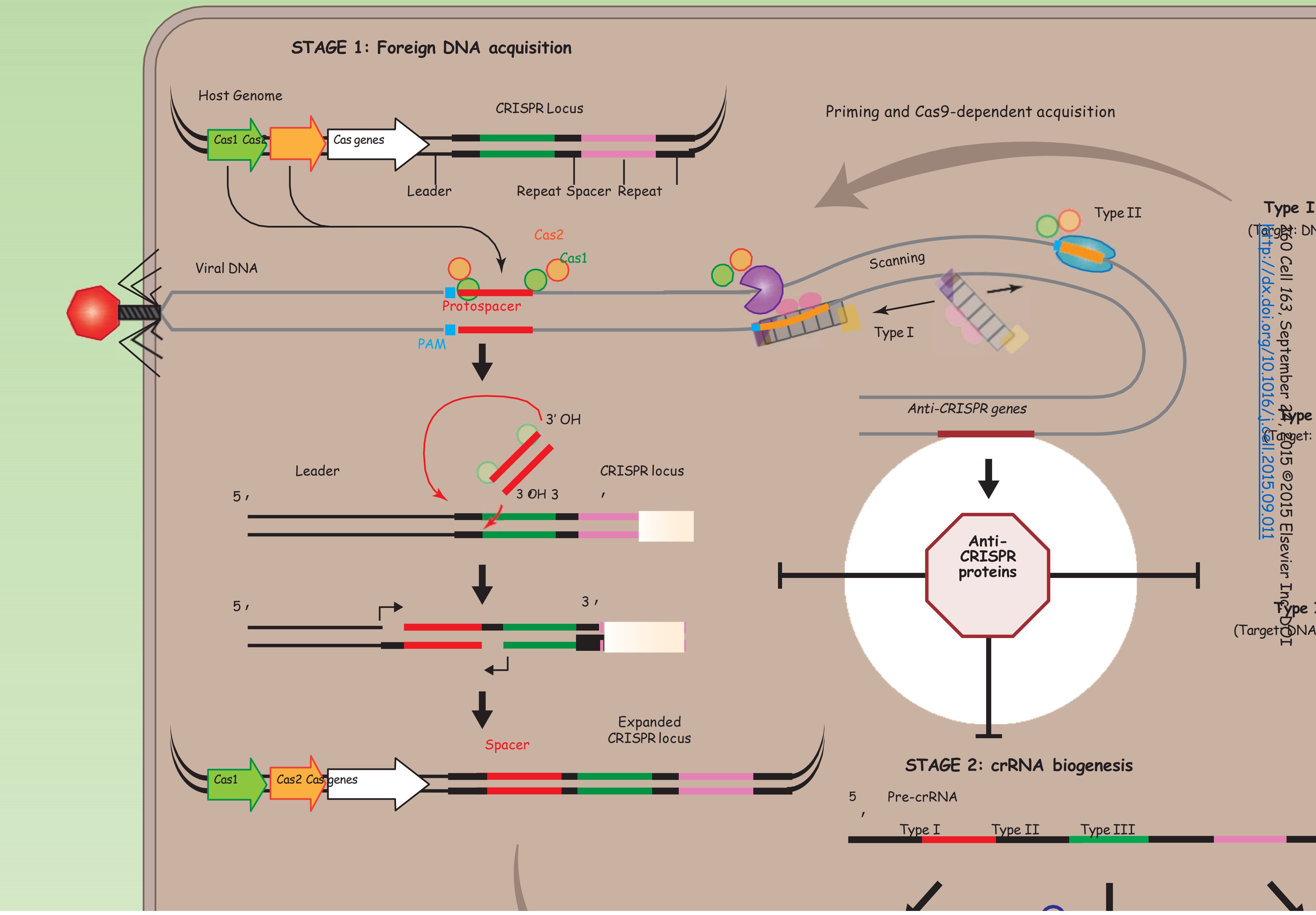
STAGE 1: Foreign DNA acquisition
Foreign nucleic acids are recognized by Cas proteins, and short fragments (30–50 base pairs) of invading DNA, called protospacers, are inserted into the host’s CRISPR locus as spacer sequences, separated by repeat sequences.
In type I and II systems, protospacers are selected from regions of invading DNA that are flanked by a 2–5 nucleotide (nt) motif called a PAM (protospacer adjacent motif) (Sorek et al., 2013).
Protospacers are generally incorporated at one end of the CRISPR locus, referred to as the leader, by a mechanism that involves Cas1, Cas2, and free 3’ hydroxyls on the protospacer (Nuñez et al., 2015).
- Protospacer integration is accompanied by the duplication of the leader-terminal repeat sequence, which may involve host polymerases and DNA repair machinery
Protospacer & PAM (sequence that being selected from invader phage genome).
Stage 2. crRNA Biogenesis
CRISPR RNA biogenesis starts with transcription, followed by processing of the primary transcript (pre-crRNA) into a library of short CRISPR-derived RNAs (crRNAs) that each contains a sequence complementary to a previously encountered foreign DNA
The crRNA-guide sequence is flanked by regions of the adjacent repeats. CRISPR RNA processing in type II systems relies on a trans-acting crRNA (tracrRNA), which contains a sequence that is complementary to the repeat sequences. These double-stranded regions are processed by RNase III while in the presence of Cas9.
In type II systems, both the tracrRNA and the crRNA are required for target interference. The two RNAs from this system have been fused into a single-guide RNA (sgRNA), and Cas9 has become a powerful tool for targeted genome engineering in a wide variety of cell types and multicellular organisms.
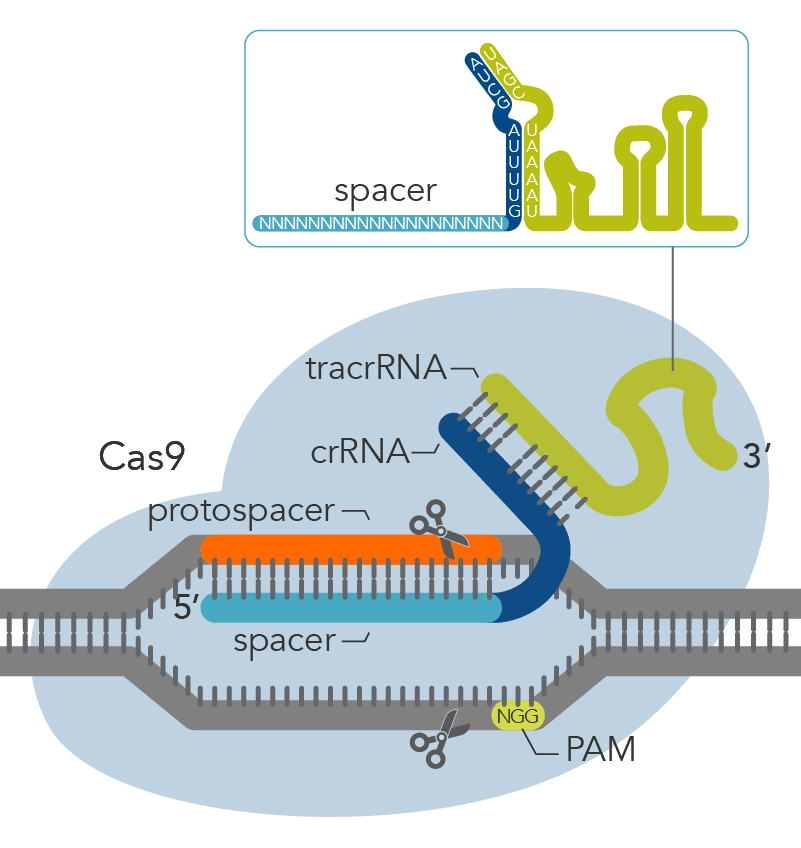
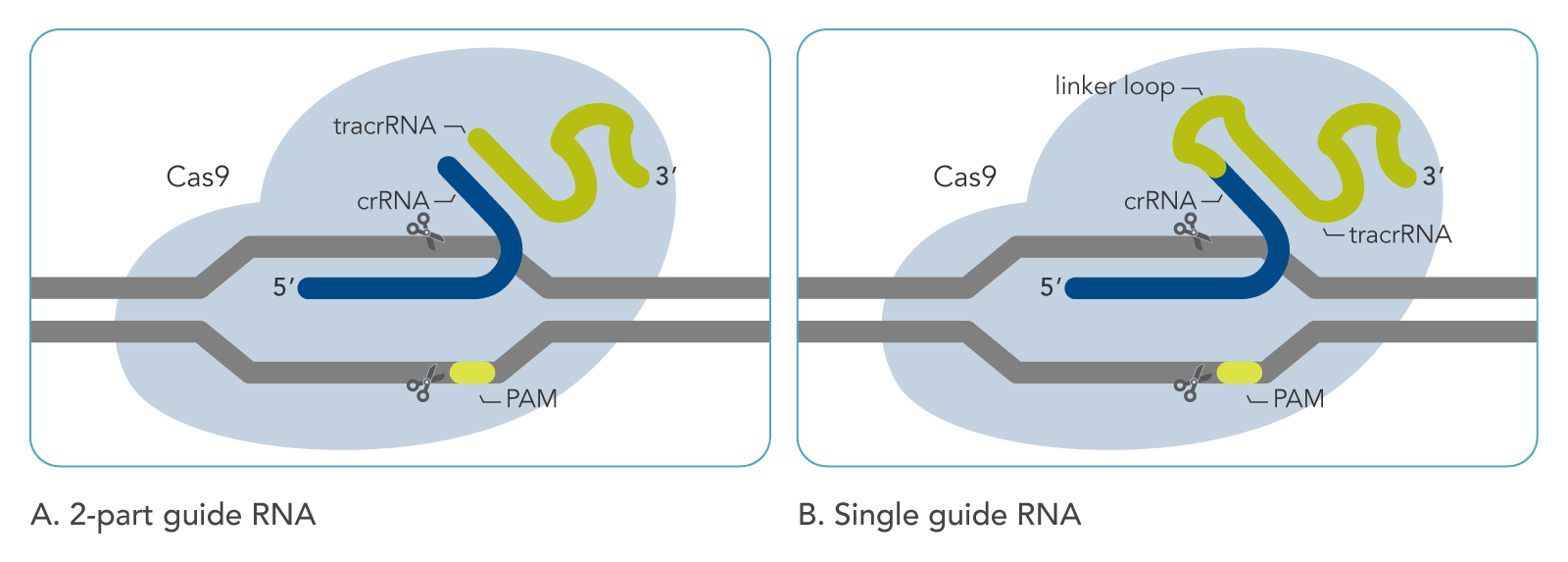
Stage 3. Target Interference
The mature crRNAs guide Cas proteins to complementary targets.
Target sequences are degraded by dedicated Cas nucleases, but the mechanisms of target degradation are diverse
Type I and II systems both target dsDNA substrates that contain a PAM and a complementary protospacer sequence.
Target cleavage in type II systems is performed by a single protein (Cas9) and two RNAs.
Type I & Type III systems rely on multi-subunit surveillance complexes that bind and cut the dsDNA substrates.
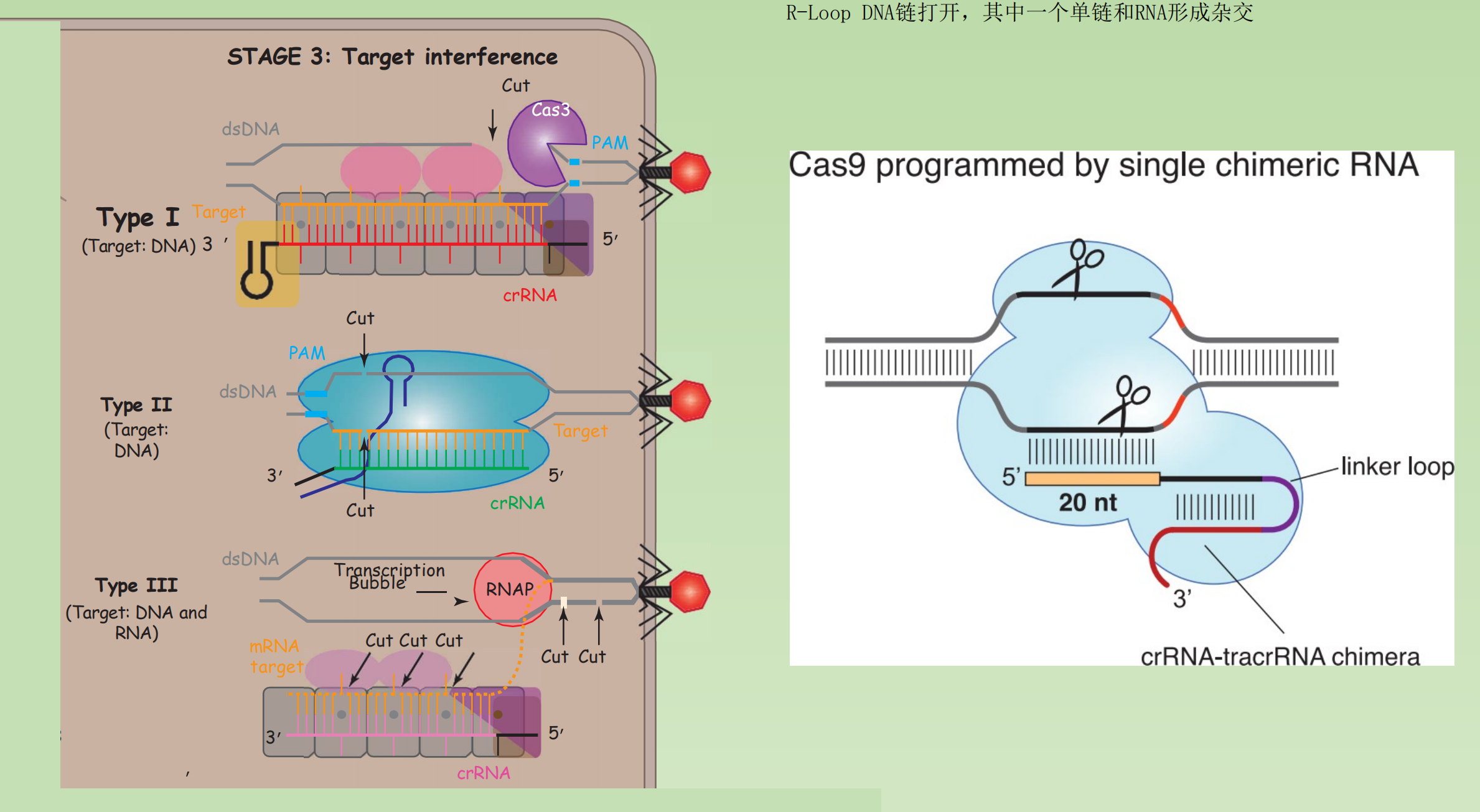
Principles
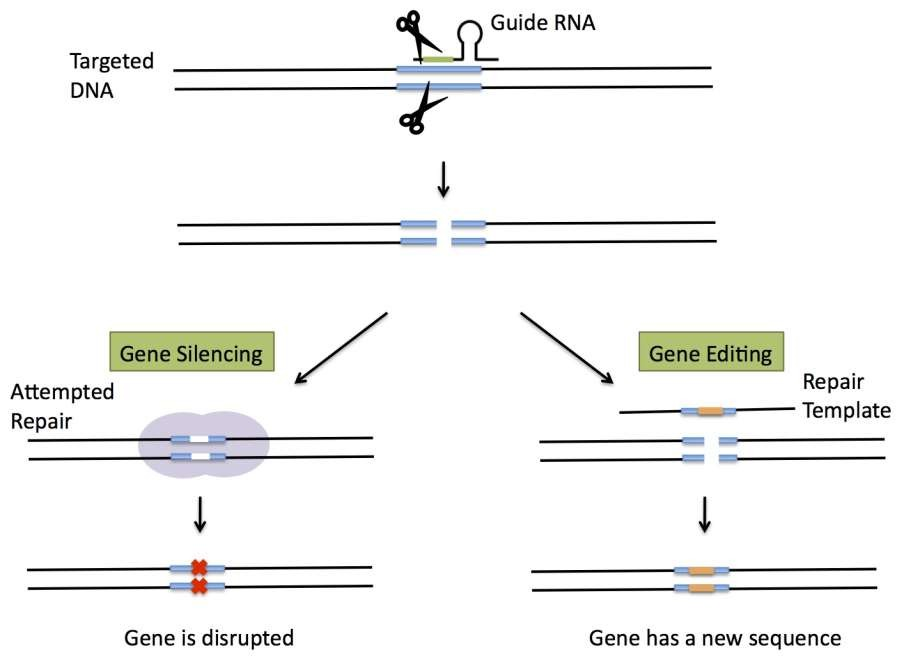
- R-loop formation between target DNA and sgRNA which is bound by Cas9
- Cas9 mediated formation of DNA double stranded break.
- DSB is healed by NHEJ pathway, accompanied with high chance of error formation (short insertion or deletion, INDELs)
- In the presence of homologous DNA, HDR mediated repair (homologous recombination) can happen.
Problem:
- High off-target rate
- High rate of unwanted mutation in the target gene.
Cas9 base editors – an example with increased precision of editing:
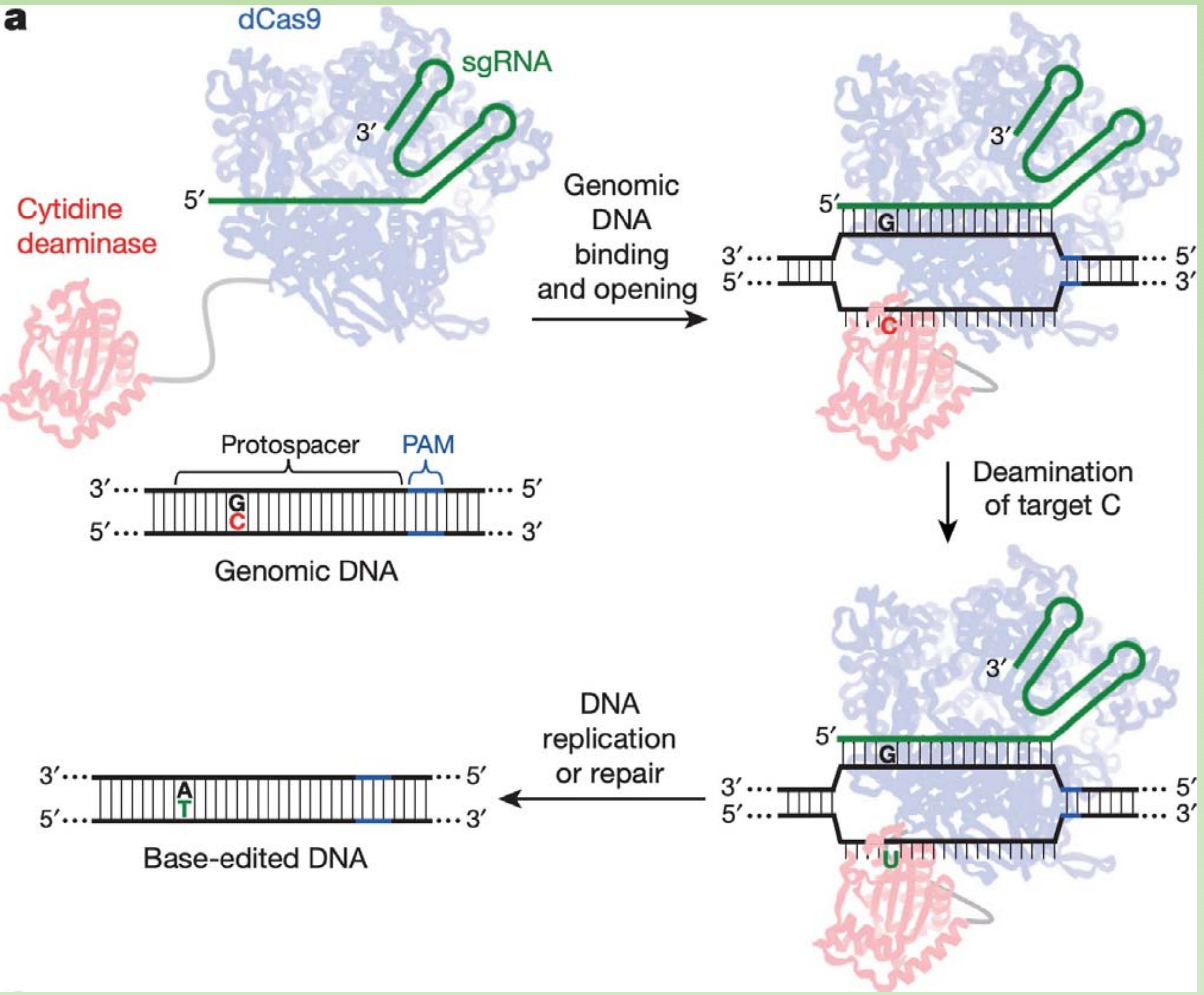
- This is the simplest version, additional elements enable increased efficiency
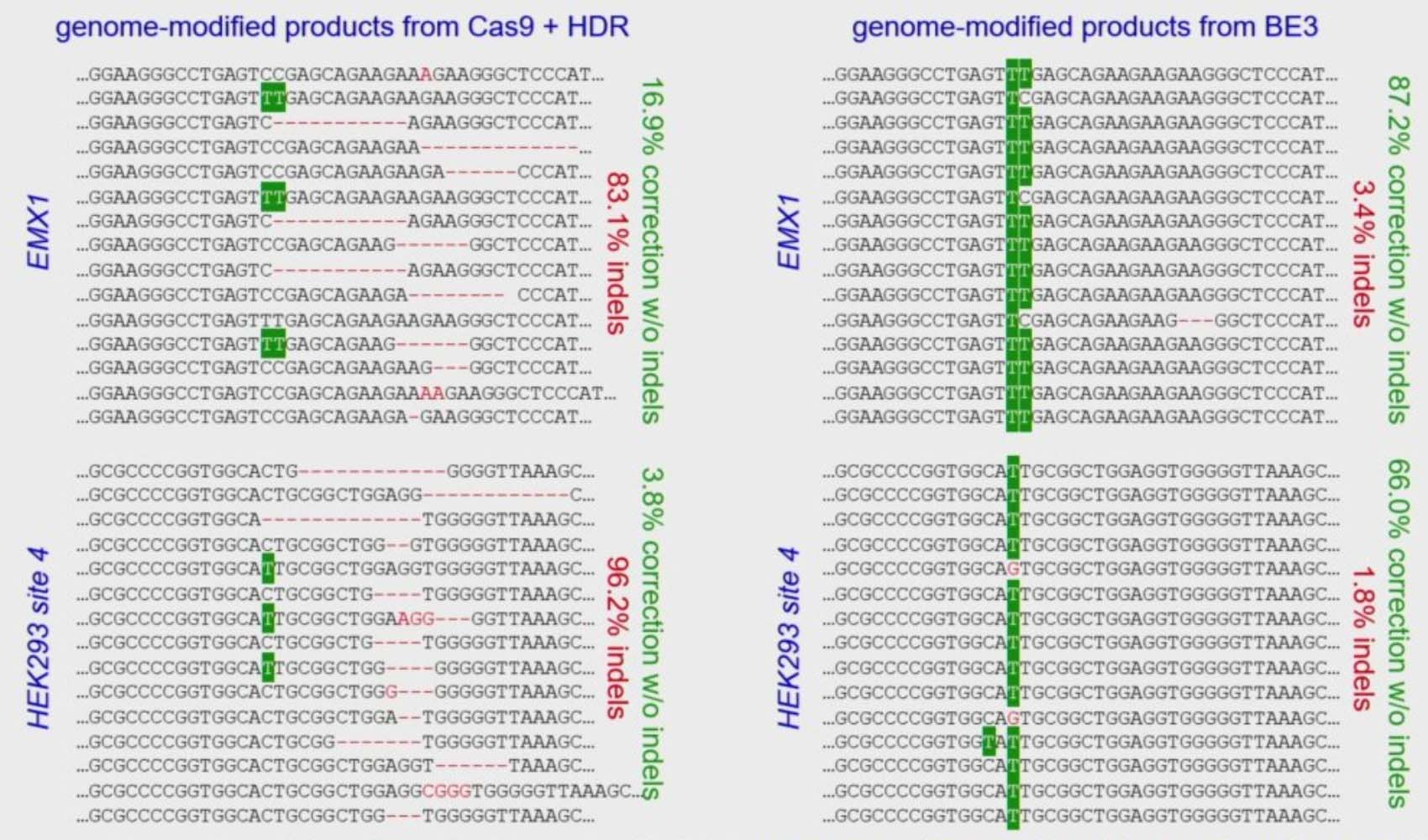
High efficiency precision editing with base editing:
二、Gene Therapy – AAV mediated Gene Therapy
Do we have the right to change ourselves ?
In 1991, James Watson declared that “many people say they are worried about the changes in our genetic instructions. But these (genetic instructions) are merely a product of evolution, shaped so we can adapt to certain conditions which might no longer exist. We all know how imperfect we are. Why not become a little better apt to survive?”
The Development of Gene Therapy
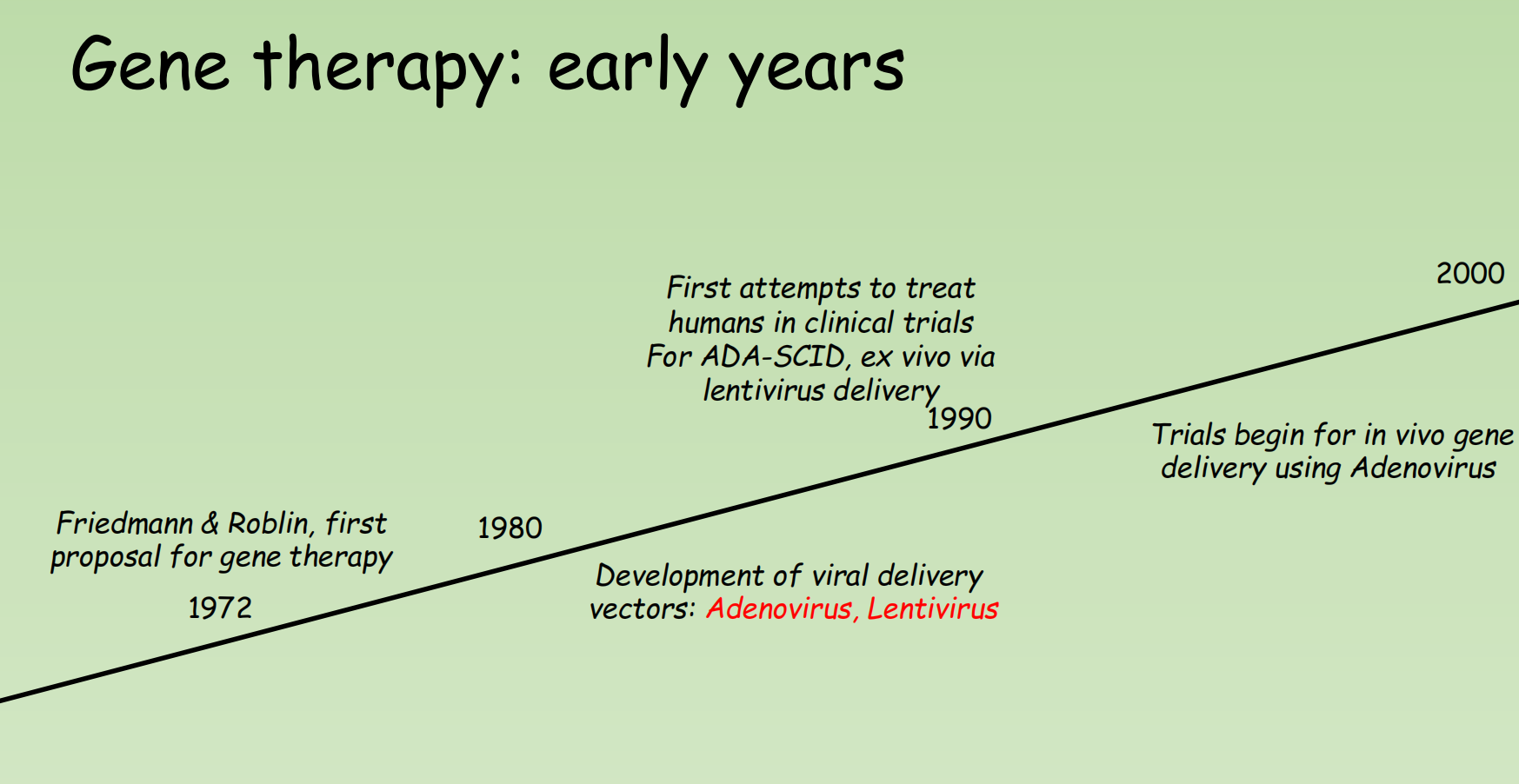
In vivo & Ex vivo:
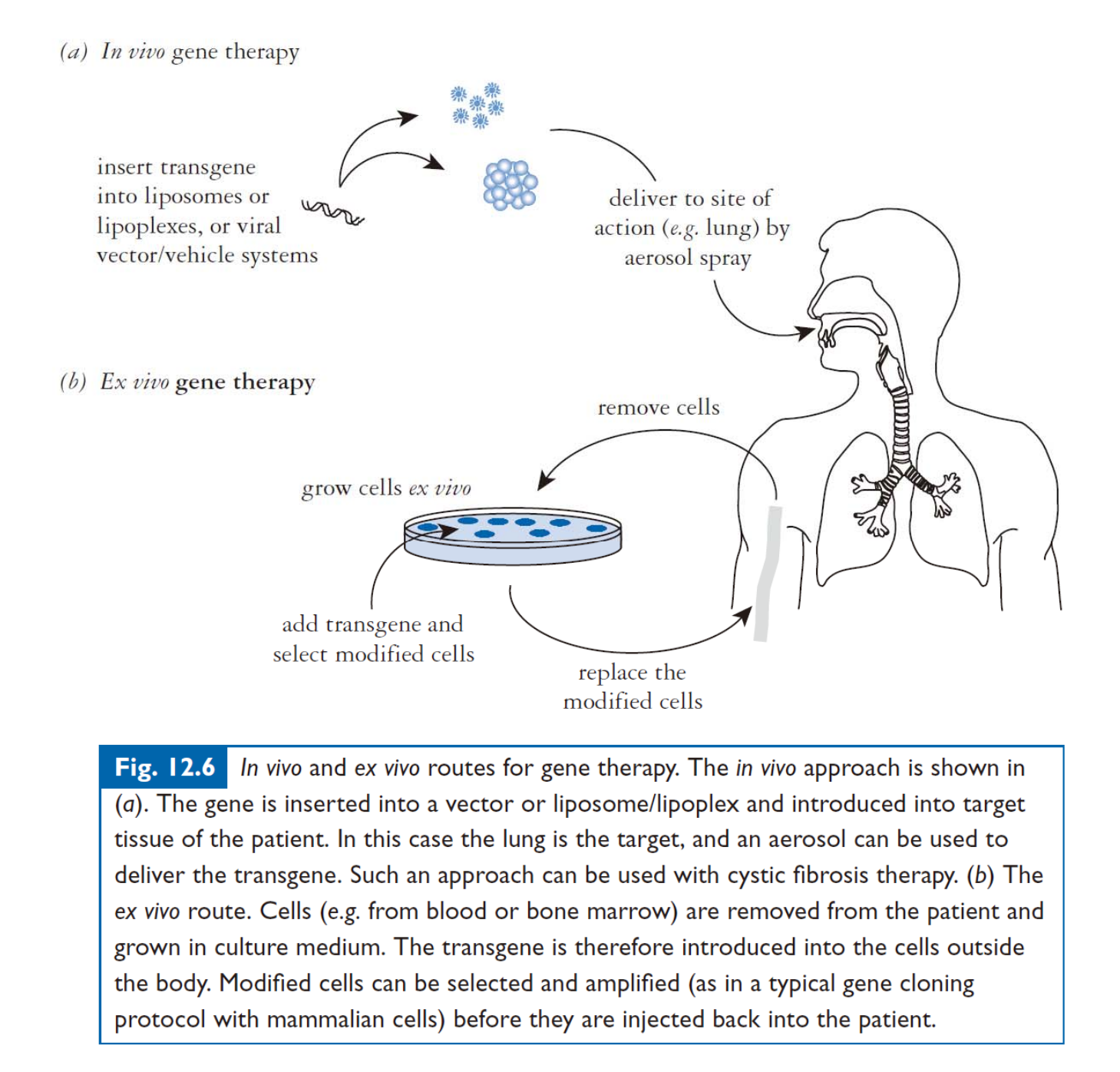
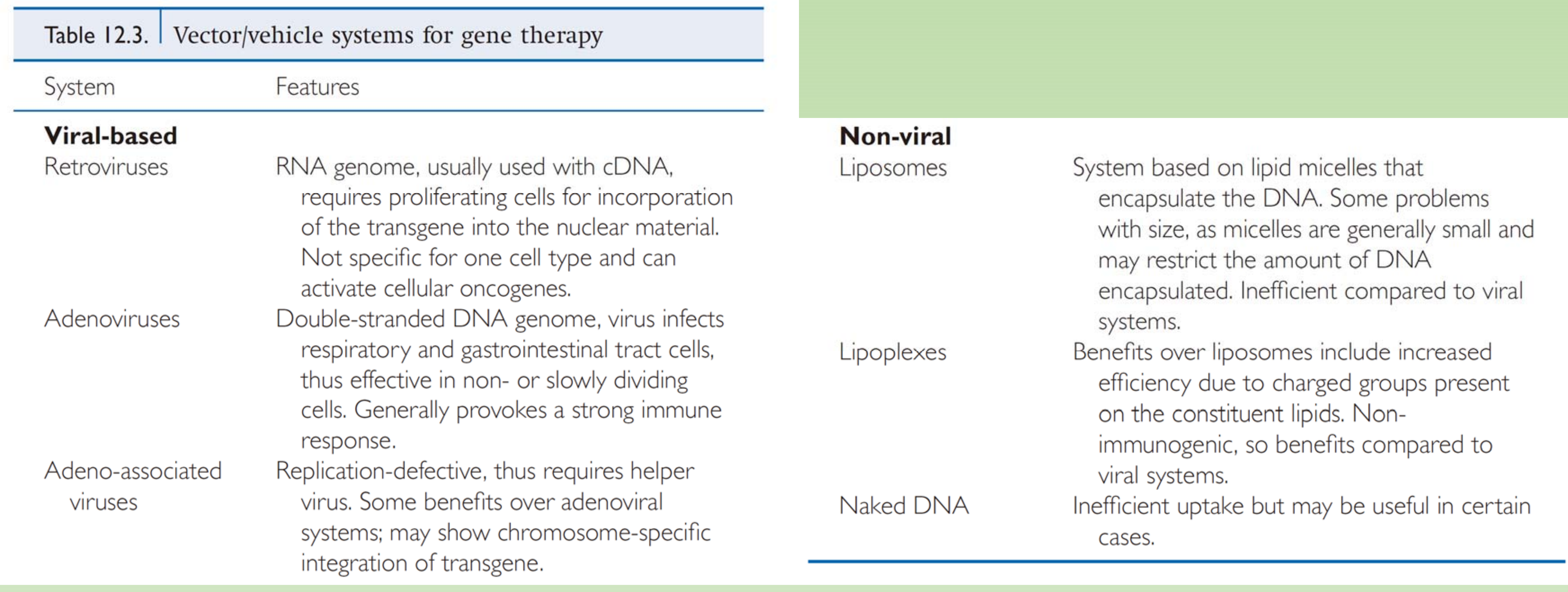
Viral-based System:
- Retrovirus
- RNA genome, usually used with cDNA. requires proliferating cells for incorporation of the transgene into the nuclear material Not specific for one cell type and can activate cellular oncogenes.
- Adenoviruses
- Double-stranded DNA genome, virus infects respiratory and gastrointestinal tract cells. thus effective in non-or slowly dividing cells, Generally provokes a strong immune response
- Adeno-associated viruses
- Replication-defective, thus requires helper virus. Some benefits over adenoviral systems: may show chromosome-specific integration of transgene
Non-viral based system:
- Liposomes
- System based on lipid micelles that encapsulate the DNA. Some problems with size, as micelles are generally small and may restrict the amount of DNA encapsulated Inefficient compared to viral systems.
- Lipoplexes
- Benefits over liposomes include increased efficiency due to charged groups present on the constituent lipids. Nonimmunogenic, so benefits compared to viral Systems
- Naked DNA
- Inefficient uptake but may be useful in certain Cases
Tragedy motivates shift toward safer delivery:

Adeno-Associated Virus (AAV)
Adeno-Associated Virus is a small non-replicative virus (dependovirus genus)
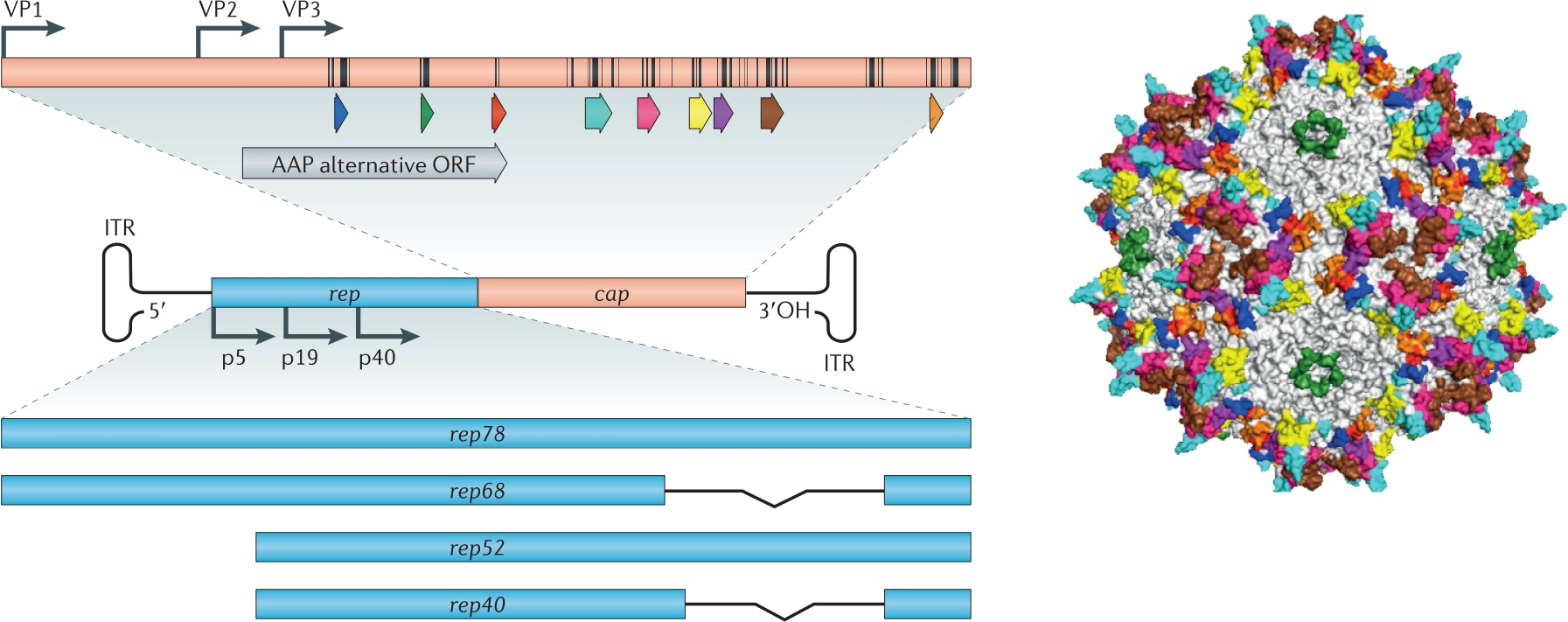
- They are small (20 nm) replication-defective, nonenveloped viruses and have linear single-stranded DNA (ssDNA) genome of approximately 4.8 kilobases (kb).
Adapting AAV for gene delivery
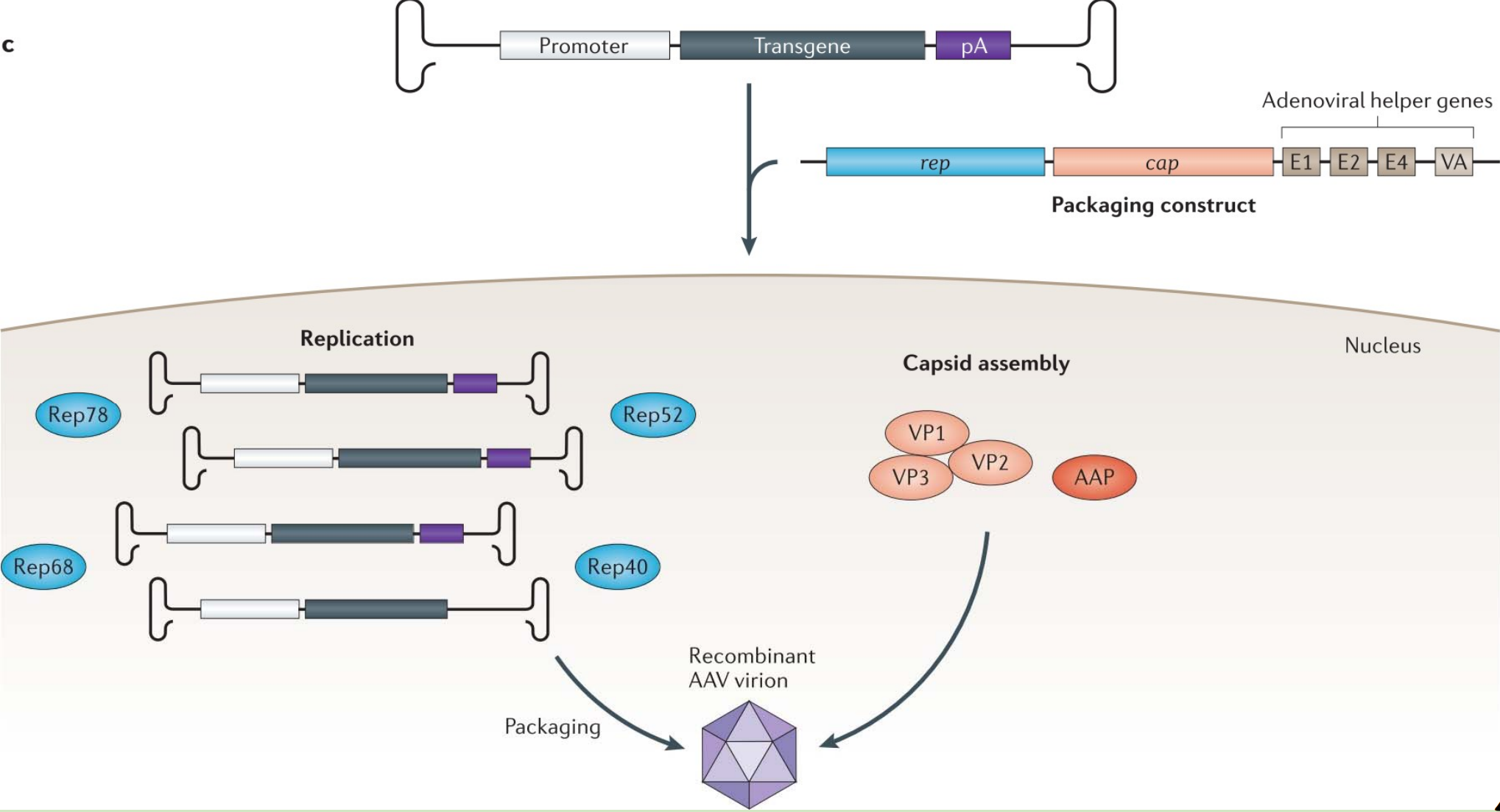
Advantages and drawbacks of using AAV in Gene therapy
• AAV is currently most promising vector for human gene therapy (In-vivo).
Benefits:
- Lack of pathogenicity and very low immunogenicity
- Can infect both dividing and non-dividing cells with high efficiency
- Has the ability to integrate into specific site – Chromosome 19.
- When used in gene therapy, the integrative capacity of AAV is often removed.
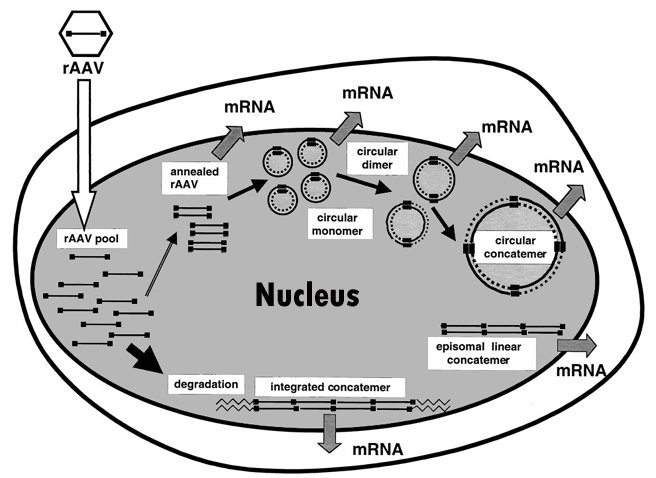
- After infection, AAV can form concatemer which can stably exist and express containing gene for the whole life cycle of the infected cell.
Drawbacks:
- The cloning capacity of the vector is relatively limited
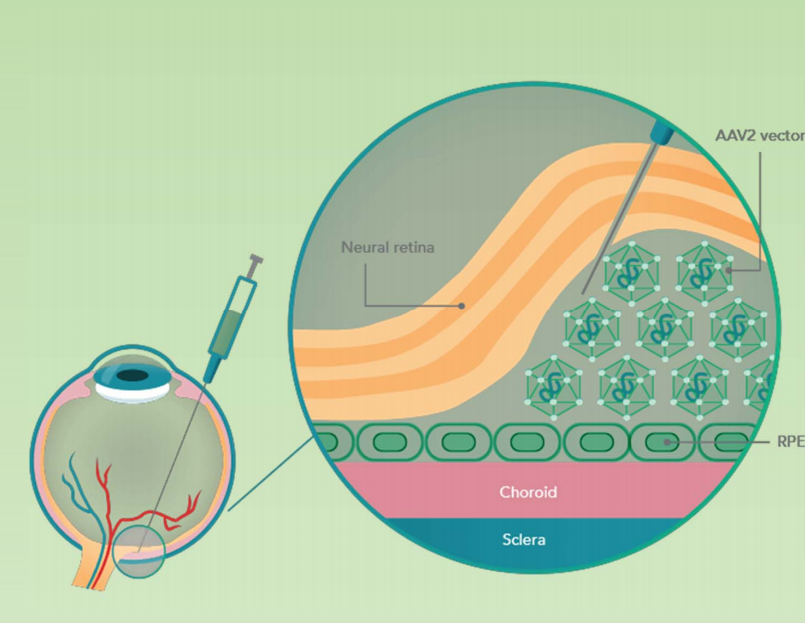
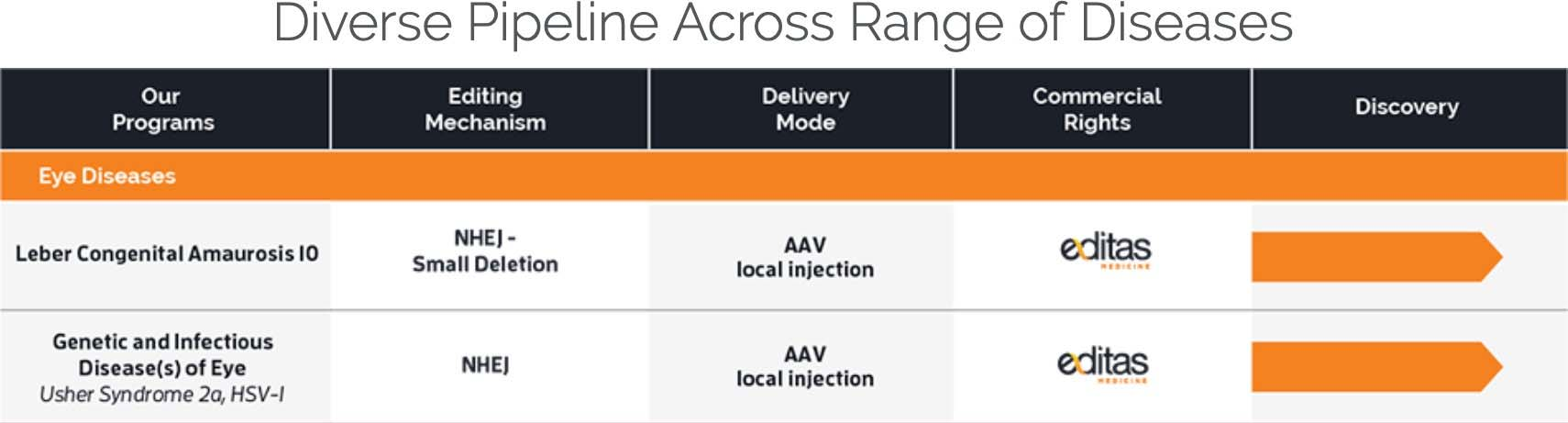
Gene therapy for Leber’s Congenital Amaurosis (LCA)
Payload: RPE65 (Retinoid somerohydrolase, makes retinol
Results in loss of vision then blindness during childhood
- Prevalence: 1 in 200.000
Subretinal AAV2 delivery
- Dose: 1.5el1vgeye in 0. 3ml rednisone 3 days prior and 7 days after iniection
- Spark Therapeutics,
- 850,000 USD per treatment
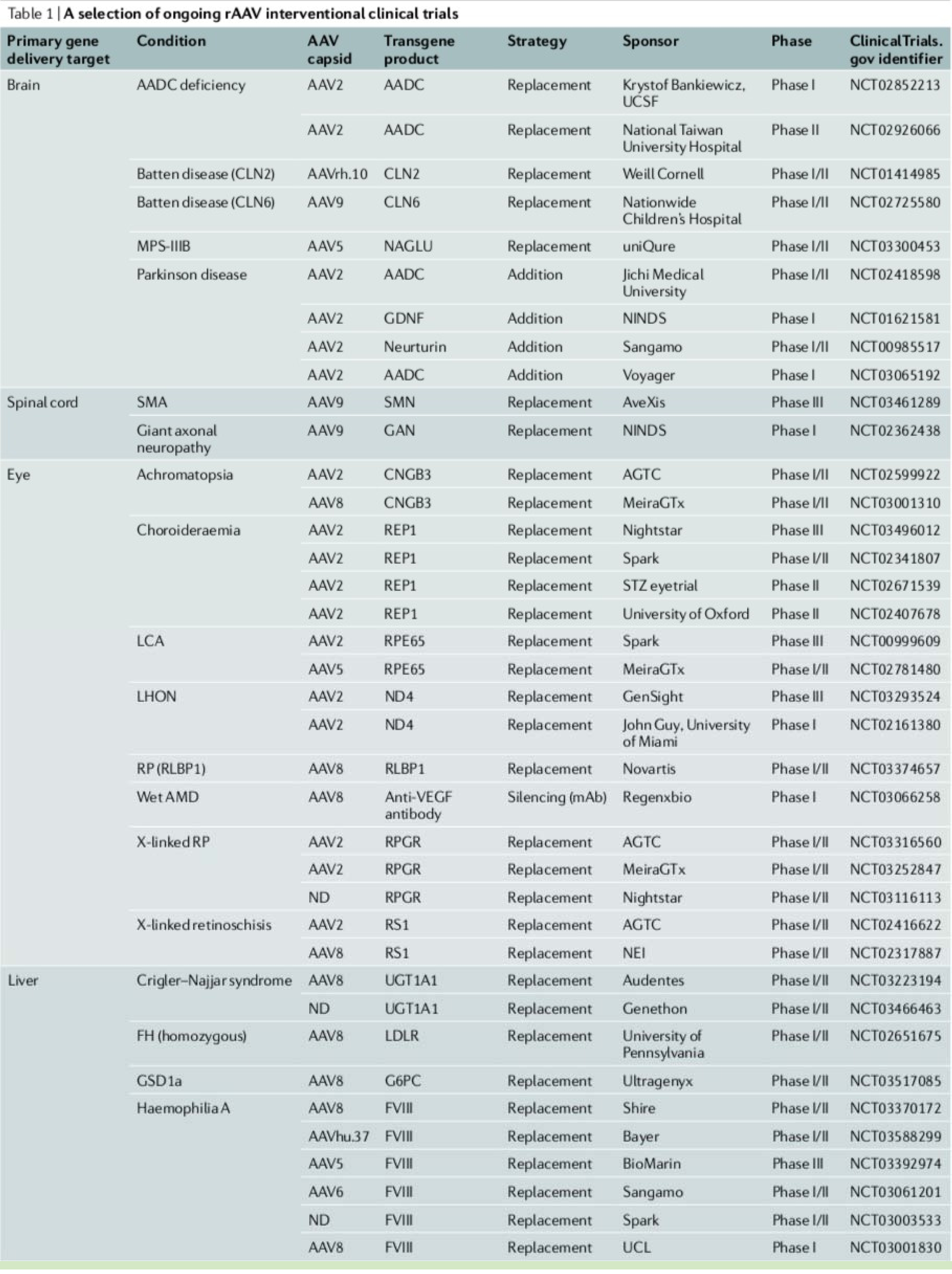
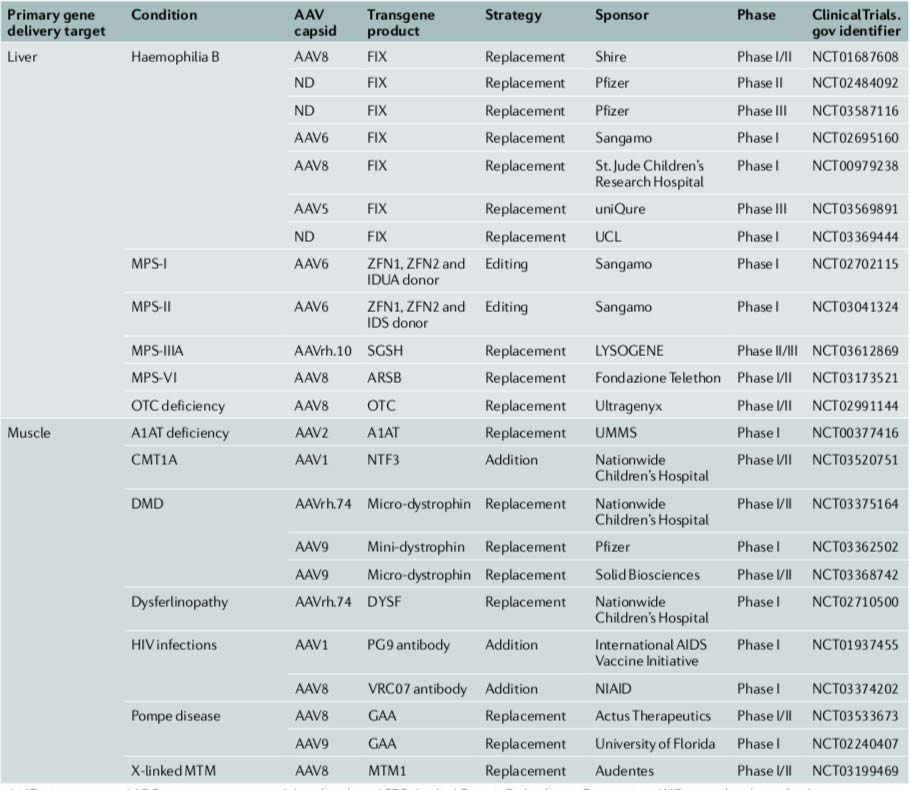
Selected AAV trials
In vivo genome editing with AAV:
SUMMARY
In vivo gene therapy delivers a genetic payload into your body’s cells
Viral capsids are efficient machines for in vivo delivery
AAV capsids enables in vivo disease treatment
- LCA, SMA, DMD, and many more…
Gene therapy field has consolidated around AAV technology
Early gene therapies are a roadmap for translation of in vivo genome editing therapies
- With innovations on payloads and delivery, curing all genetic diseases is within reach in our lifetime