L1&2 Intro and Genetic Structure
一、Introduction to Genetic Engineering
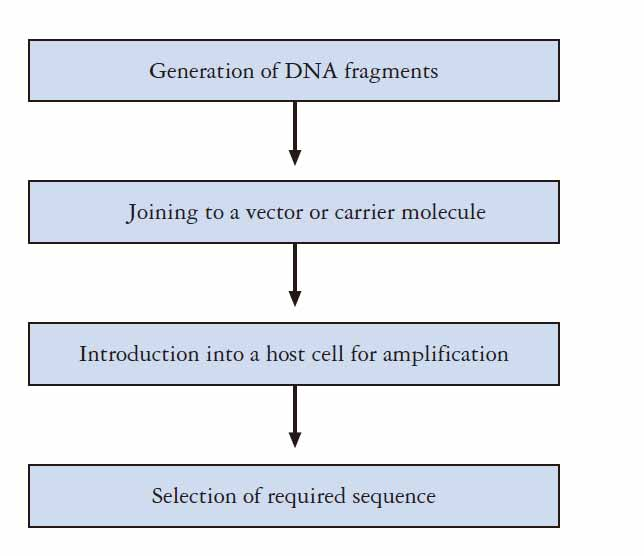
Genetic engineering is a term which involves the group of techniques used to cut up and join together genetic material, especially DNA from different biological species, and to introduce the resulting hybrid DNA into an organism in order to form new combinations of heritable genetic material.
- It is a recombinant DNA technique
- It involves transfer the recombinant DNA into a host cell–transformation
- The newly transformed recombinant DNA is heritable
Important Discoveries in Genetic Engineering
Gregor Mendel – basic principle of genetics. Law of segregation and Law of independent assortment. Knowing the basis of genetics.
Thomas Hunt Morgan – established the chromosome theory of heredity. He showed that genes are linked in a series on chromosomes and are responsible for identifiable, hereditary traits. He adopted the term gene, which was introduced by the Danish botanist Wilhelm Johannsen in 1909. Mechanism of Mendelian Heredity (1915). Morgan’s work played a key role in establishing the field of genetics. He received the Nobel Prize for Physiology or Medicine in 1933.
James Watson and Francis Crick – The discovery of DNA structure 1953
Theoretical foundation of Genetic Engineering
The discovery made by Gregor Mendel, Thomas Hunt Morgan, James Watson and Francis Crick pretty much laid the theoretical foundation for Genetic Engineering.
- Development of various enzymes which are important tools of Genetic Engineering
- Development of carriers of recombinant DNA – Vectors
- Development of methods for delivering the recombinant DNA into the host cell. – Transformation
1. The general procedure of genetic engineering
2. A timeline of the history of Genetic Engineering
DNA polymerase - A. Kornberg in 1956 purified DNA polymerase.
DNA ligase - In 1967 the enzyme DNA ligase was isolated by Weiss and Richardson.
Restriction Enzyme - isolation of the first restriction enzyme in 1970.
Recombinant DNA molecules – the first pieces were generated at Stanford University in 1972, utilizing the cleavage properties of restriction enzymes (scissors) and the ability of DNA ligase to join DNA strands together (glue). (By P. Berg)
Vector - The key to gene cloning is to tended in 1973 by joining DNA fragments to the plasmid pSC101, ensure that the target sequence is replicated in a suitable host cell. which is an extrachromosomal element isolated from the bacterium Escherichia coli. (By Cohen and Boyer)
DNA sequencing - 1977, Sanger DNA sequencing
PCR - 1987, K.B. Mullis developed PCR.
The objectives of Genetic Engineering
- Bioreactor
- Prokaryotic or Eukaryotic cells are cultured as host cells for foreign
gene expression, to produce useful proteins in low cost - Mainly used for pharmaceutical purpose !
- advantages genetically engineered drug:
- Not limited by the slaughter of animals.
- Large quantities can be made quickly.
- No risk of transferring infections.
- More effective at treating disease as animal proteins is different to human proteins.
- No ethical issues concerning the use of animals.
- Prokaryotic or Eukaryotic cells are cultured as host cells for foreign
- Aid basic research
- Example: Reporter genes can be used to measure promoter activity or tissue-specific expression.
Summary of objectives of genetic engineering:
- Basic Research, Gene’s function, link phenotype to genotype
- Production of useful proteins and compound – Bioreactor or Molecular Farming.
- Generation of transgenic plants and animals
- Medical diagnosis and treatment
- Genome analysis by DNA sequencing
- Synthetic Biology
- Forensic Analysis; (法医分析)
- Paternity Disputes;
二、Basic Knowledge for Genetic Engineering
Important knowledges:
- The difference between GE and Molecular Biology
- The Central Dogma of Molecular Biology
- DNA and RNA structure, DNA replication
- Mutations
- Gene structure and organization
- Genome organization
The difference between GE and Molecular Biology
Molecular Biology instruct you knowledge around the central dogma, you get to know how a cell works at molecular level in a nutshell
The knowledge you learned in the course of Molecular Biology are generated mostly (if not all)from experimental works
Genetic Engineering pretty much tells you how do those experiments work
This course is best served as a companion for your daily lab works
The central dogma
The Flow of Information: DNA to RNA to protein
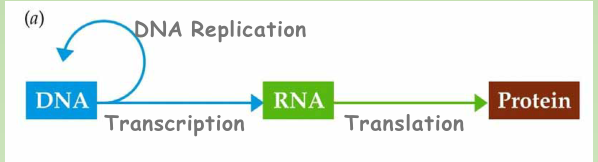
A gene is expressed in two
- steps: DNA is transcribed to RNA
- Then RNA is translated into protein.
Modification of Central Dogma:
- RNA dependent RNA polymerase (RDR)
- RNA dependent DNA polymerase (Reverse transcriptase)
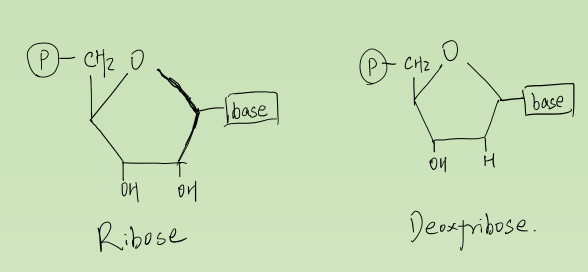
Structure of DNA and RNA
1. Deoxyribonucleic Acid and Ribonucleic Acid
Made up of nucleotides (DNA molecule) in a DNA double helix.
- ~2 nm wide
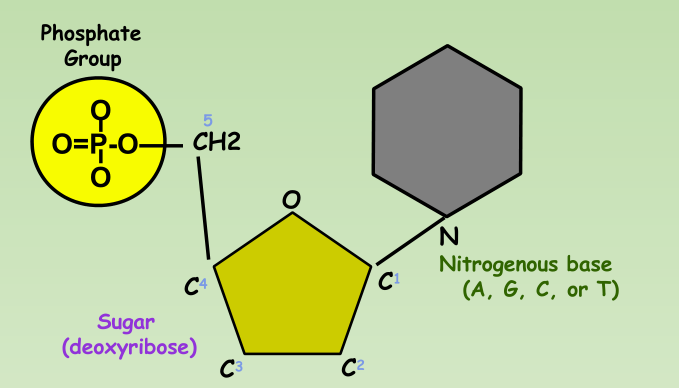
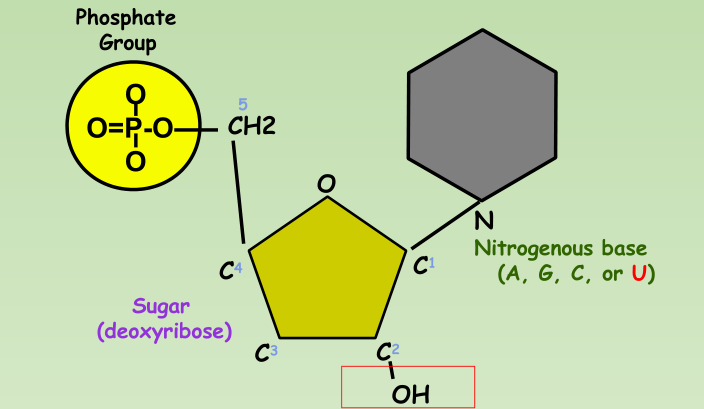
Nucleotide:
- Phosphate group
- 5-carbon sugar
- Nitrogenous base
DNA Nucleotide:
RNA Nucleotide:
DNA Replication
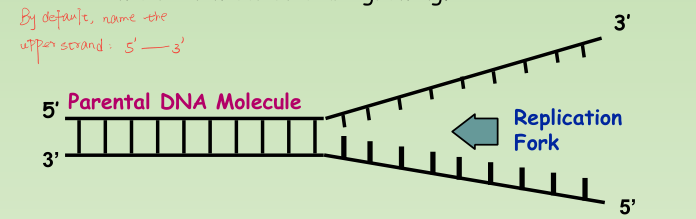
Origins of replication
- Replication Forks: hundreds of Y-shaped regions of replicating DNA molecules where new strands are growing.
- By default, name the upper strand as 5’ $\rarr$ 3’
- Origins of replication: Replication Bubbles:
- Hundreds of replicating bubbles (Eukaryotes).
- Single replication fork (bacteria).
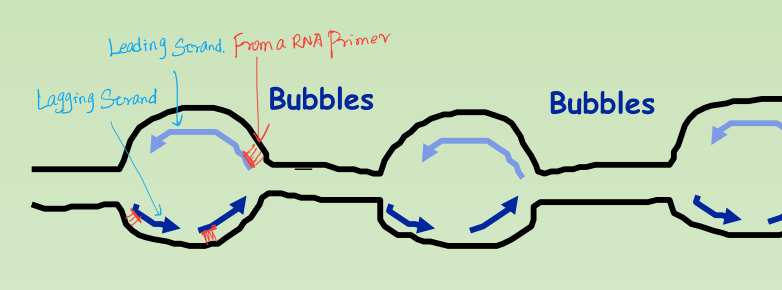
1. Synthesis of the new DNA Strands:
- Leading Strand: synthesized as a single polymer in the 5’ to 3’ direction.
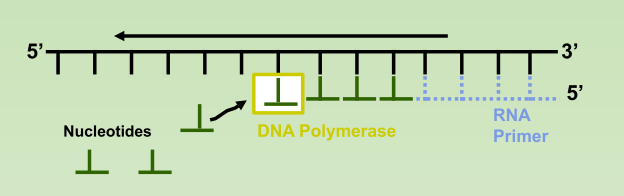
- Lagging Strand: also synthesized in the 5’ to 3’ direction, but discontinuously against overall direction of replication.
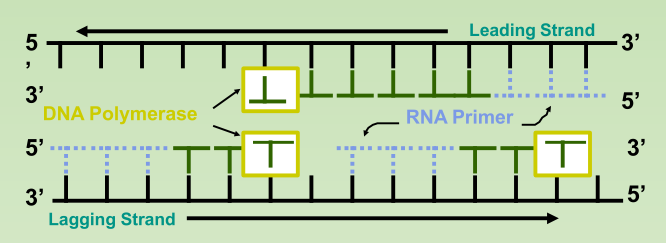
- Okazaki Fragments: series of short segments on the lagging strand.
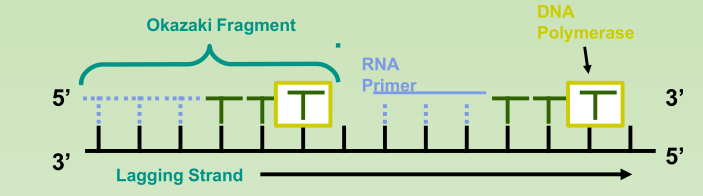
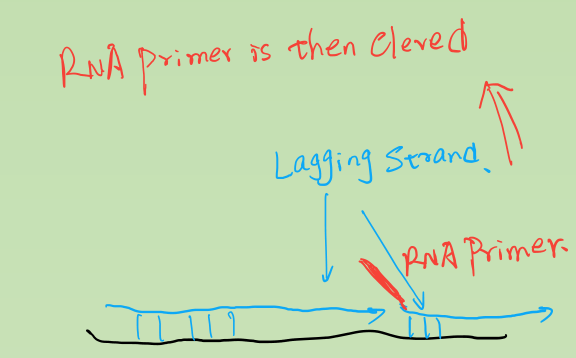
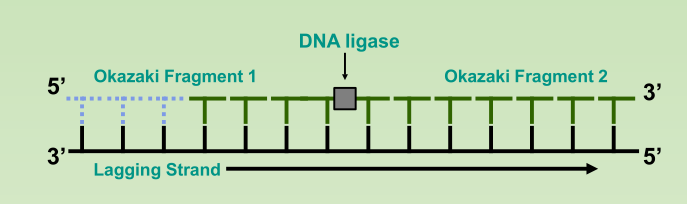
- DNA ligase: a linking enzyme that catalyzes the formation of a covalent bond from the 3’ to 5’ end of joining stands.
- Example: joining two Okazaki fragments together
Gene structure and organization
1. Prokaryotic gene structure
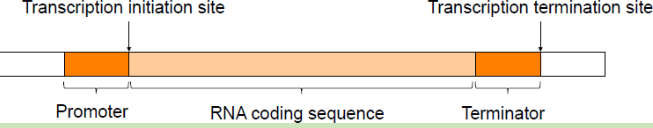
Prokaryotic Gene is composed of three regions:
- Promoter region
- RNA coding sequence
- Terminator region
Prokaryotic gene is continues and uninterested where there is no introns present
The region 5’ of the promoter sequence is called upstream sequence and the region 3’ of the terminator sequence is called downstream sequence.
Promoter region
This is situated on upstream of the sequence that codes for RNA.
This is the site that interact RNA polymerase before RNA synthesis(Transcription).
Promoter region provides the location and direction to initiate transcription
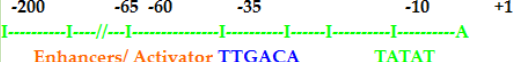
- At -10 there is a sequence TATAAT or PRIBNOW BOX.
- At -35 another consensus sequence TTGACA
- These two are the most important promoter elements recognized by transcription factors.
RNA coding sequence
The DNA sequence that will become copied into an RNA molecule (RNA transcript).
Starts with an initiator codon and ends with termination codon
- No introns (uninterrupted).
- Collinear to its mRNA
Any nucleotide present on the left is denoted by (-)symbol and the region is called upstream element. E.g. -10,-20,-35 etc.
Any sequence to the right of the start is downstream elements and numbered as +10,+35 etc.).
Terminator region
The region that signal the RNA polymerase to stop transcription from DNA template.
Transcription termination occur through Rho dependent or Rho independent manner
2. Eukaryotic gene structure
Eukaryotic gene are complex structures compared that prokaryotic gene.
They are composed of following regions:
- Exons
- Introns
- Promoter sequences
- Terminator sequences
- Upstream sequences
- Downstream sequences
- Enhancers and silencers (upstream or downstream)
- Signals (Upstream sequence signal for addition of cap. Downstream sequences signal for addition of poly A tail.)
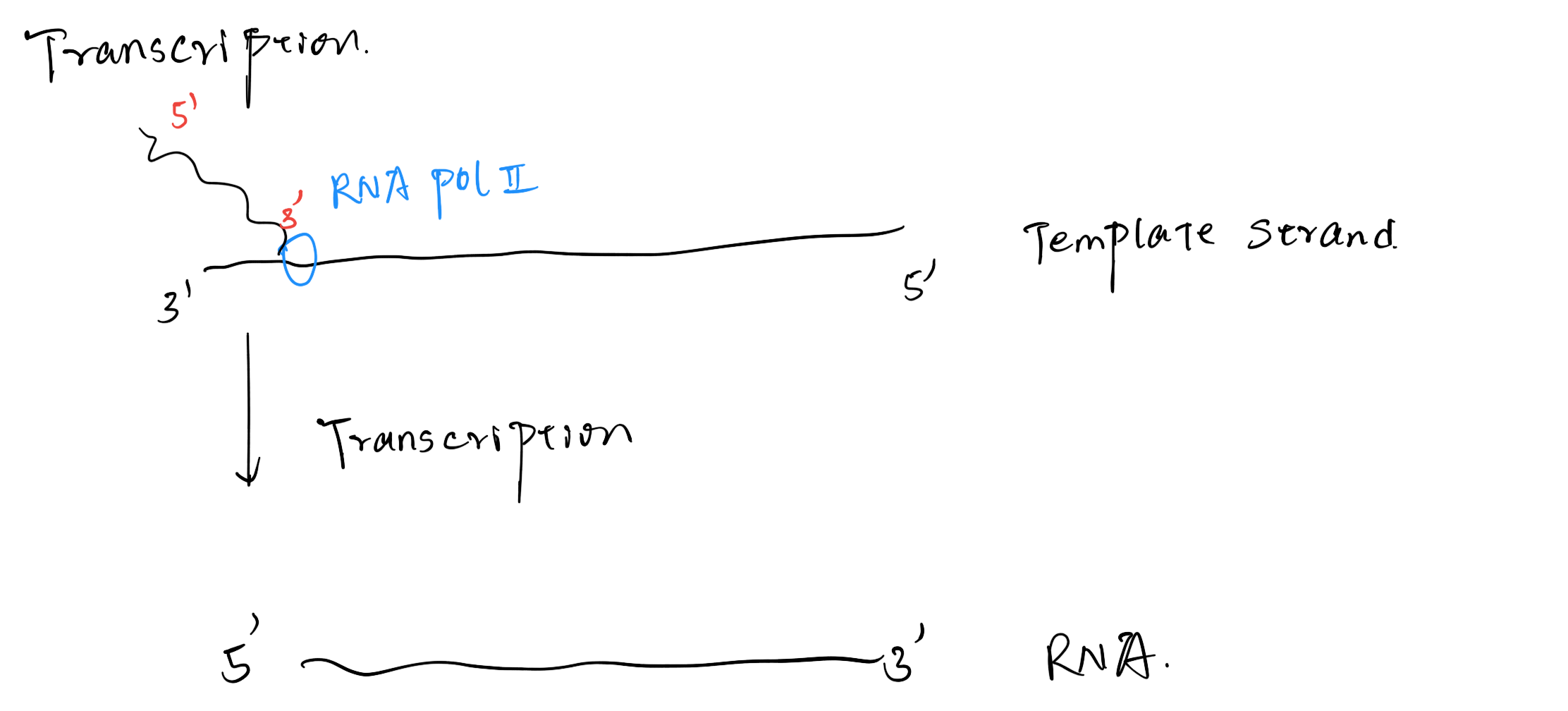
Different Strands
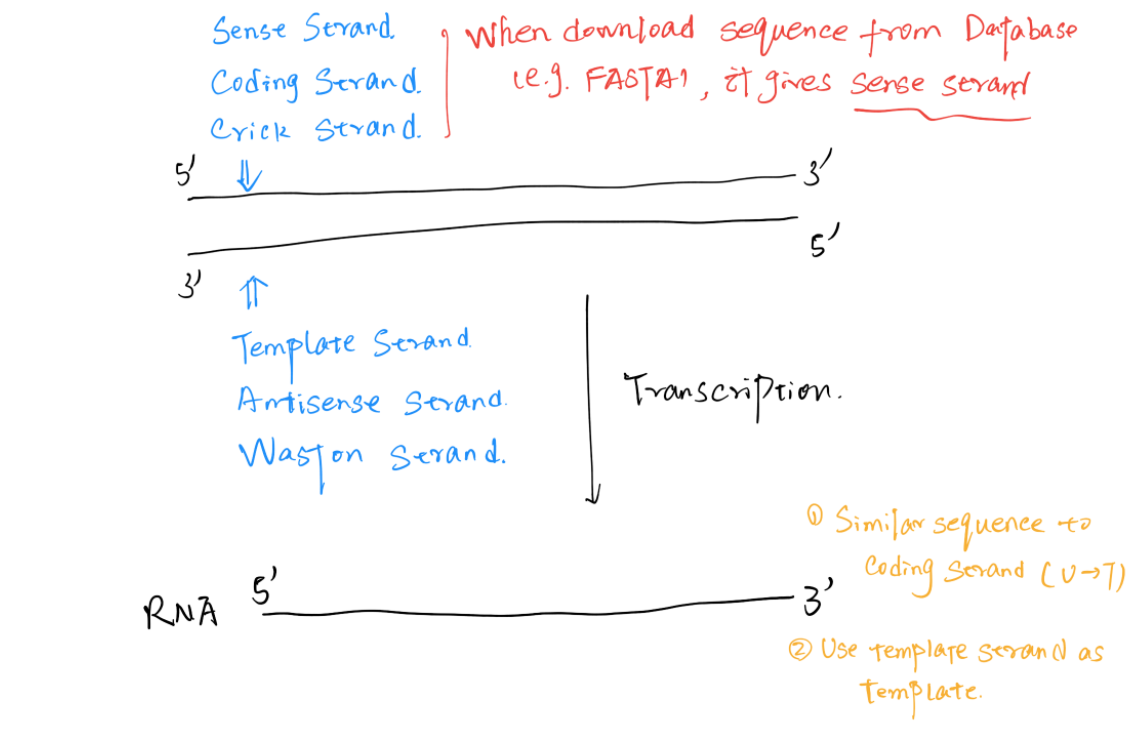
The RNA transcripted has similar sequence to the Coding Strand (Sense Strand, Crick Strand)
- The DNA sequence downloaded from database are always coding strand
The RNA Pol use the Template Strand (Antisense Strand, Waston Strand) to synthesize RNA
The Coding Region Structure
(1) General Structure
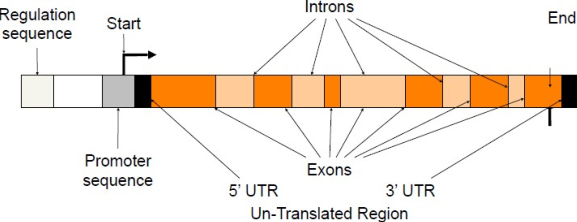
- TSS:transcription start site
- TES/TTS: transcription termination site
- PAS: polyadenylation site
- Enhancers/silencers
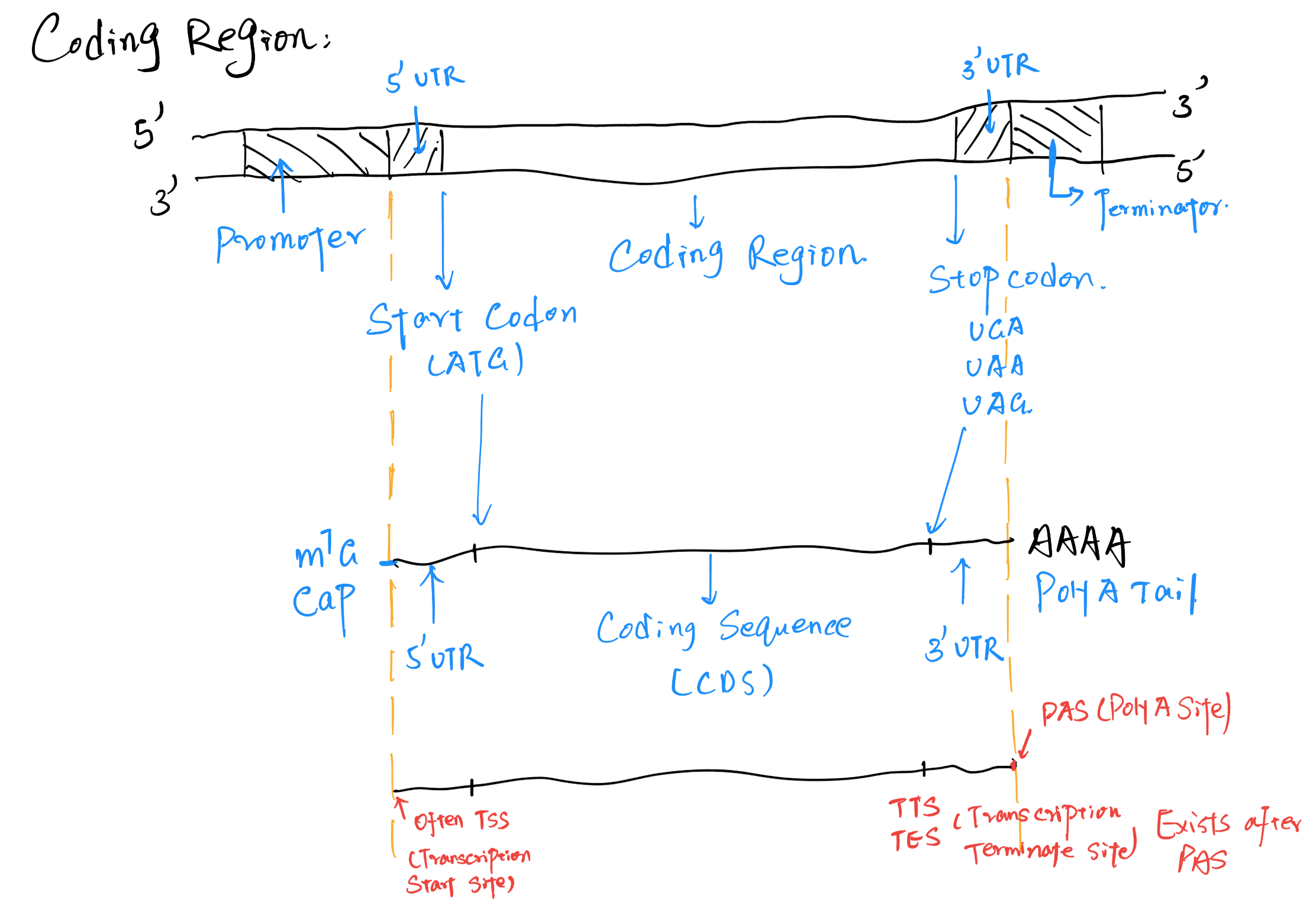
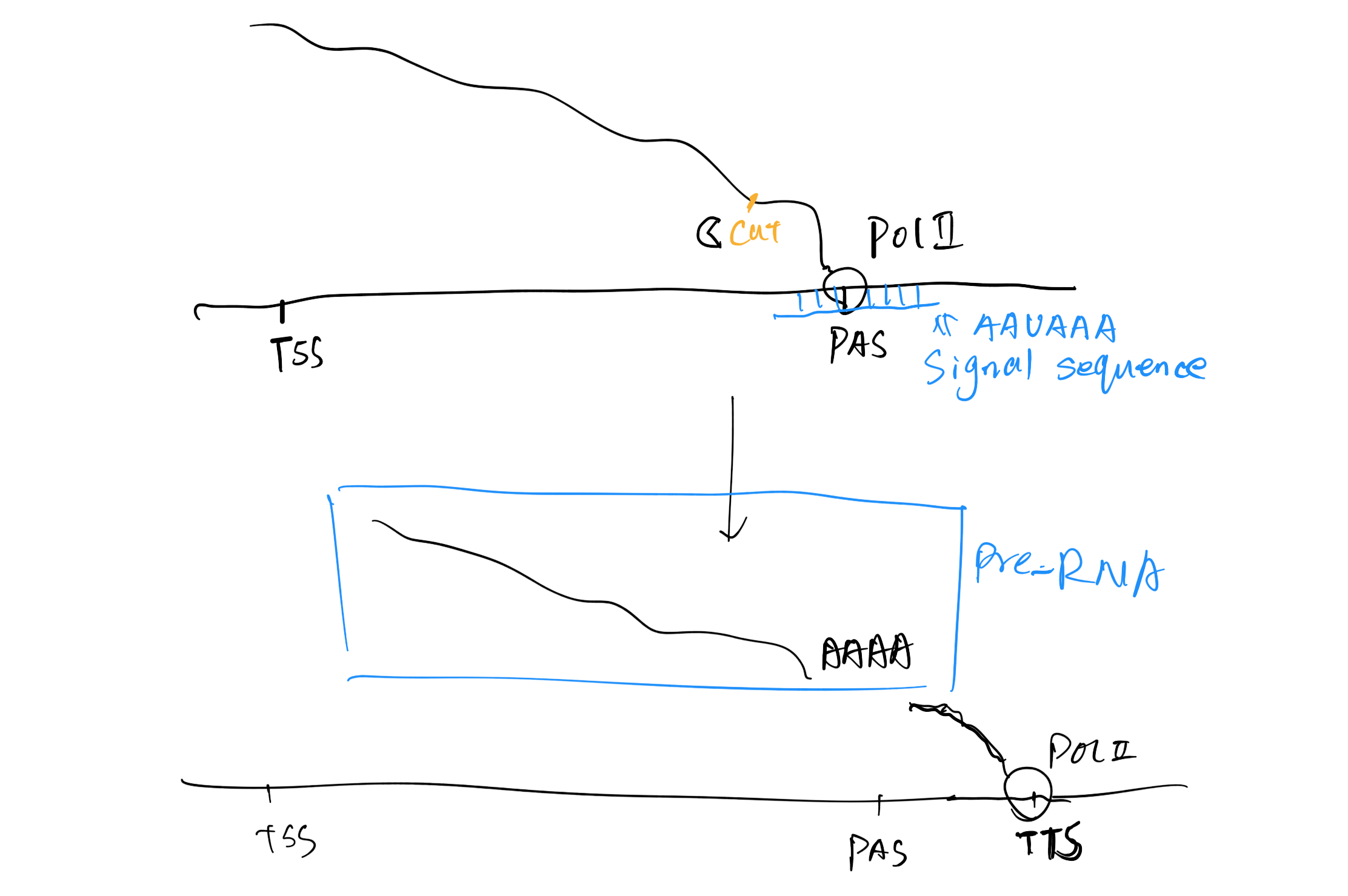
(2) Introns and Exons
Exons
- Coding sequence, transcribed and translated.
- Coding for amino acids in the polypeptide chain.
- Vary in number ,sequence and length. A gene starts and ends with exons.(5’to 3’).
- Some exon includes untranslated(UTR) region.
Introns
- Coding sequences are separated by noncoding sequences called introns.
- They are removed when the primary transcript is processed to give the mature RNA
- All introns share the base sequence GT in the 5’end and AG in the 3’end.
- Introns were 1st discovered in 1977 independently by Phillip Sharp and Richard Roberts.
Significance of Introns
Introns don’t specify the synthesis of proteins but have other important cellular activities.
Many introns encodes RNA’s that are major regulators of gene expression.
Contain regulatory sequences that control trancription and mRNA processing.
Introns allow exons to be joined in different combinations(alternative splicing),resulting in the synthesis of different proteins from the same gene.
Important role in evolution by facilitating recombination between exons of different genes(exon shuffling)
(3) Promoters
A promoter is a regulatory region of DNA located upstream controlling gene expression.
- Core promoter – transcription start site(-34) Binding site for RNA polymerase
and it is a general transcription factor binding sites - Proximal promoter-contain. primary regulatory element.
- These together are responsible for binding of RNA polymerase II which is responsible for transcription
(4) Upstream (5’end)
5’UTR serve several functions including mRNA transport and initiation of translation.
Signal for addition of cap(7 methyl guanisine) to the 5’end of the mRNA.
The cap facilitates the initiation of translation
Stabilization of mRNA.
(5) Downstream (3’end)
3’UTR serves to add mRNA
Stability and attachment site for poly-A-tail.
The translation termination codon TAA.
AATAA sequence signal for addition of poly A tail
(6) Terminator
Recognized by RNA polymerase as a signal to stop transcription
(7) Enhancer
Enhances the transcription of a gene upto few thousand bp upstream
- Enhancer influence the 3D structure of DNA molecule, therefore enhance transcription
- 使得DNA链上两个区域空间位置更加接近
(8) Silencers
Reduce or shut down the expression of a near by gene.
3. Salient features of gene
Number of genes in each organism is more than the number of chromosomes; hence several genes are located on each chromosome.
The genes are arranged in a single linear order like beads on a string.
Each gene occupies specific position called locus.
If the position of gene changes, character changes.
Genes can be transmitted from parent to off springs.
Genes may exist in several alternate formed called alleles.
Genes are capable of combined together or can be replicated during a cell division.
Genes may undergo for sudden changes in position and composition called mutation.
Genes are capable of self duplication producing their own exact copies
4. Mutations
Mutation- change in DNA sequence leading to a different protein sequence being produced
- same codon produced.
Missense mutation - different codon introduced
- Partially acceptable
Nonsense mutation -stop codon introduced
- Usually unacceptable
Silent Mutation – change of DNA sequence without a subsequent change in the amino acid
Genome organization
Nucleosome – The basic structural subunit of chromatin, consisting of ~200 bp of DNA and an octamer of histone proteins.
10 nm fiber – A linear array of nucleosomes generated by unfolding from the natural condition of chromatin.
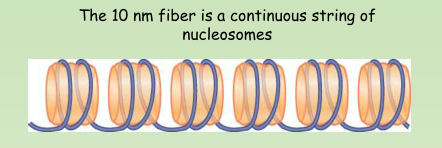
Histone tails – Flexible amino or carboxy-terminal regions of the core histones that extend beyond the surface of the nucleosome.
- Histone tails are sites of extensive posttranslational modification
Nonhistone – Any structural protein found in a chromosome except one of the histones
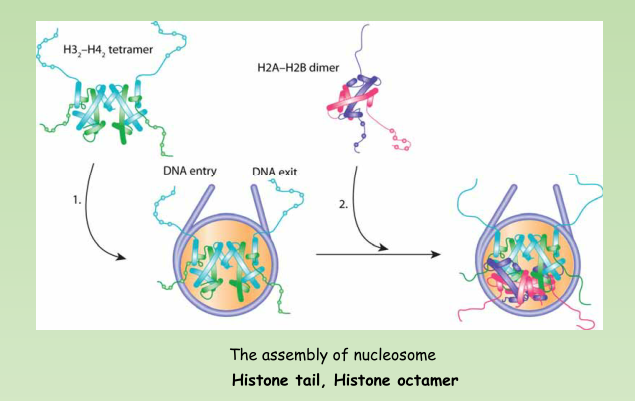
- A nucleosome contains ~200 bp of DNA and two copies of each core histone (H2A, H2B, H3, and H4).
- The histone octamer has a structure of an H32-H42 tetramer associated with two H2A-H2B dimers
30 nm fiber – A coil of nucleosomes
- It is the basic level of organization of nucleosomes in chromatin.
1. Histone Modification
Nucleosomes Are Covalently Modified – mainly at histone tails
Histones are modified by methylation, acetylation, phosphorylation, and other modifications
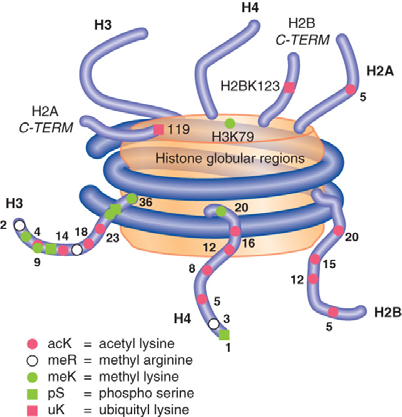
Histones are modified by methylation, acetylation,phosphorylation, and other modifications.
Combinations of specific histone modifications define the function of local regions of chromatin; this is known as the histone code.
Histone modification can affect process such as transcription and DNA replication.
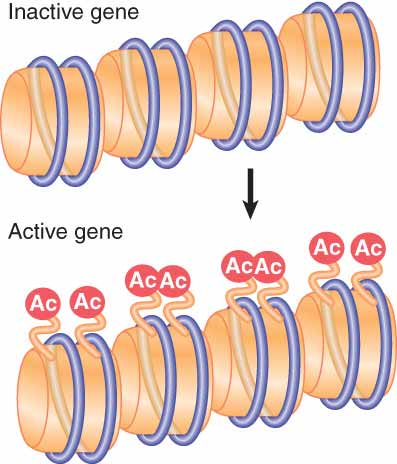
Acetylation associated with gene activation occurs by directly modifying specific sites on histones that are already incorporated into nucleosomes.
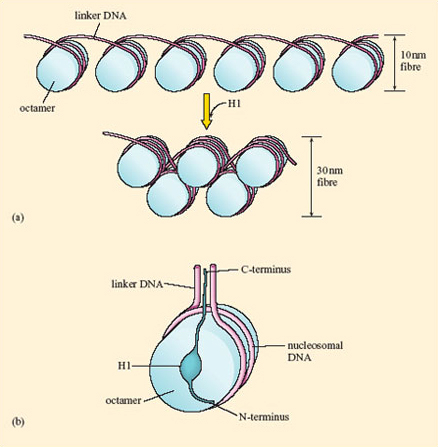
- Linker histone H1 helped the package of nucleosome into 30nm fiber
- Affect heterochromatin
- H1 is not essential in eukaryotes