L21 Apoptosis
一、Programmed cell death
In most cases, this programmed cell death occurs by a process called apoptosis
Cells dying by apoptosis undergo characteristic morphological changes.
- They shrink and condense,
- the cytoskeleton collapses
- the nuclear envelope disassebles
- the nuclear chromatin condenses and breaks up into fragments
- The cell surface often bulges (凸起;膨胀) outward and, if the cell is large, it breaks up into membrane-enclosed fragments called apoptotic bodies
Two distinct types of cell death: necrosis vs apoptosis
In contrast to apoptosis, animal cells that die in response to an acute insult, such as trauma or a lack of blood supply, usually do so by a process called cell necrosis. Necrotic cells swell and burst, spilling their contents over their neighbors and eliciting an inflammatory response
In most cases, necrosis is likely to be caused by energy depletion, which leads to metabolic defects and loss of the ionic gradients that normally exist across the cell membrane.
- One form of necrosis, called necroptosis, is a form of programmed cell death that is triggered by a specific regulatory signal from other cells, although we are only just beginning to understand the underlying mechanisms.
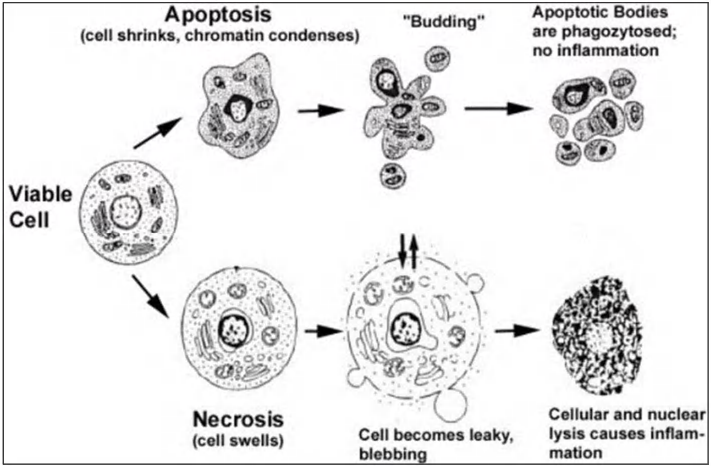
Different types of death: necrosis vs apoptosis
- Necrosis: cells swell and burst, content spillage (溢出;溢出量), can cause inflammation
- Apoptosis: cells die “clean and tidy” (eaten and digested by neighboring cells or macrophages)
- Apoptosis - is the original term introduced by Kerr et al. to define a type of cell death with specific morphological features.
- Term used to describe “Programmed Cell Death” originally by Kerr, J. F., Wyllie, A. H. & Currie, A. R. Apoptosis: a basic biological phenomenon with wideranging implications in tissue kinetics. Br. J. Cancer 26, 239-257 (1972).
Apoptosis is NOT a synonym of programmed cell death or caspase activation
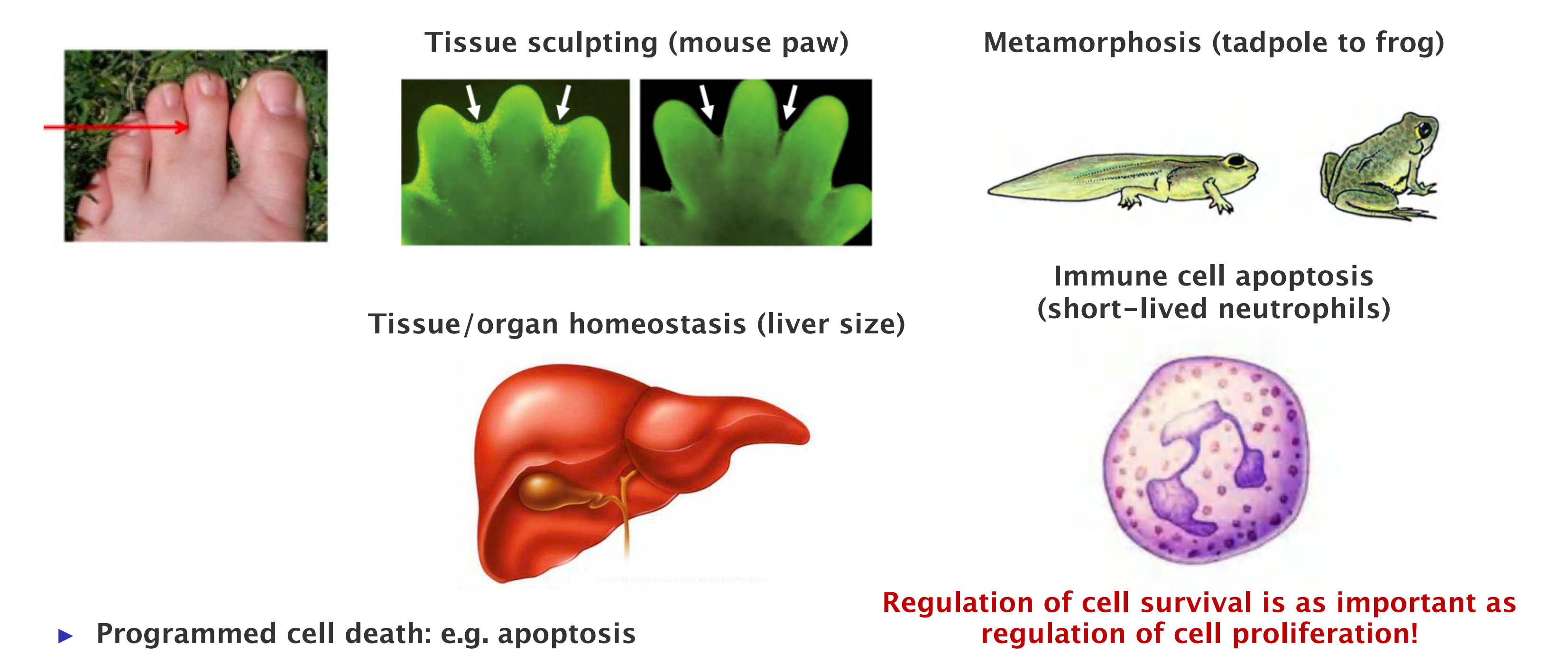
- Programmed cell death: e.g. apoptosis
- It is a natural part of organism development and maintenance.
- Eliminating cells that are abnormal, misplaced, nonfunctional, or potentially dangerous
Apoptosis
During apoptosis, cells undergo morphological changes
Characteristics of apoptosis:
- Cell shrinkage and chromatin condensation
- PS flipping to outside
- DNA fragmentation
- Nuclear membrane disruption
- Cytoskeleton collapses
- Cell surface bleb ⏤ apoptotic bodies
1. Understanding apoptosis
C. elegans as new model organism to analyze organ development
Brenner established C. elegans as a novel experimental model organism.
This provided a unique opportunity to link genetic analysis to cell division, differentiation and organ development – and to follow these processes under the microscope.
Cell death is part of the normal differentiation process
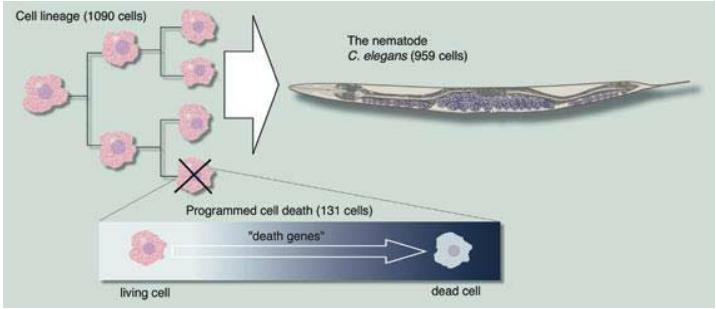
Sulston mapped a cell lineage where every cell division and differentiation could be followed in the development of a tissue in C. elegans.
He showed that specific cells undergo programmed cell death as an integral part of the normal differentiation process.
He also identified the first mutation of a gene participating in the cell death process.
Cell death is controlled by genes
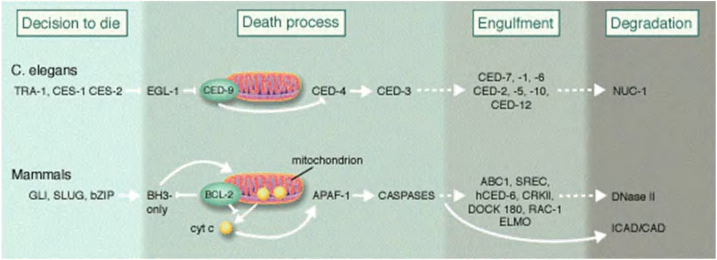
Horvitz discovered and characterized key genes controlling cell death in C. elegans.
He has shown how these genes interact with each other in the cell death process and that corresponding genes exist in humans.
二、The underlying mechanisms of apoptosis
Players in apoptosis
Apoptosis Depends on an Intracellular Proteolytic Cascade That Is Mediated by Caspases
Apoptosis is triggered by members of a family of specialized intracellular proteases, which cleave specific sequences in numerous proteins inside the cell
- These proteases have a cysteine (半胱氨酸蛋白酶) at their active site and cleave their target proteins at specific aspartic acids; they are therefore called caspases (c for cysteine and asp for aspartic acid).
There are two major classes of apoptotic caspases: initiator caspases and executioner caspases.
Caspases
Extrinsic pathway
Intrinsic pathway
Bcl-2
Inhibitor of apoptosis (IAPs)
Caspases
Apoptosis depends on an intracellular proteolytic cascade
Proteolysis is catalyzed by caspases.
Caspases have a cysteine in their active site and cleave their target proteins at specific aspartic acids (→ caspase)
Caspases are zymogens (synthesized as inactive precursors), → procaspase
- “pre-” was adopted as the prefix that means “before N-terminal cleavage during a secretion event”
- “pro-“ was adopted to describe the protein prior to this proteolytic processing event.

Apoptosis is triggered by a cascading reaction of initiator and executioner caspases
Two types of caspases:
- initiator caspases
- executioner caspases
Initiator caspases, as their name implies, begin the apoptotic process.
- They normally exist as inactive, soluble monomers in the cytosol.
- An apoptotic signal triggers the assembly of large protein platforms that bring multiple initiator caspases together into large complexes. Within these complexes, pairs of caspases associate to form dimers, resulting in protease activation
The major function of the initiator caspases is to activate the executioner caspases
- These normally exist as inactive dimers. When they are cleaved by an initiator caspase at a site in the protease domain, the active site is rearranged from an inactive to an active conformation.
- One initiator caspase complex can activate many executioner caspases, resulting in an amplifying proteolytic cascade.
Various experimental approaches have led to the identification of over a thousand proteins that are cleaved by caspases during apoptosis.
- These include the nuclear lamins, the cleavage of which causes the irreversible breakdown of the nuclear lamina
Signal-mediated cascading activation of apoptosis
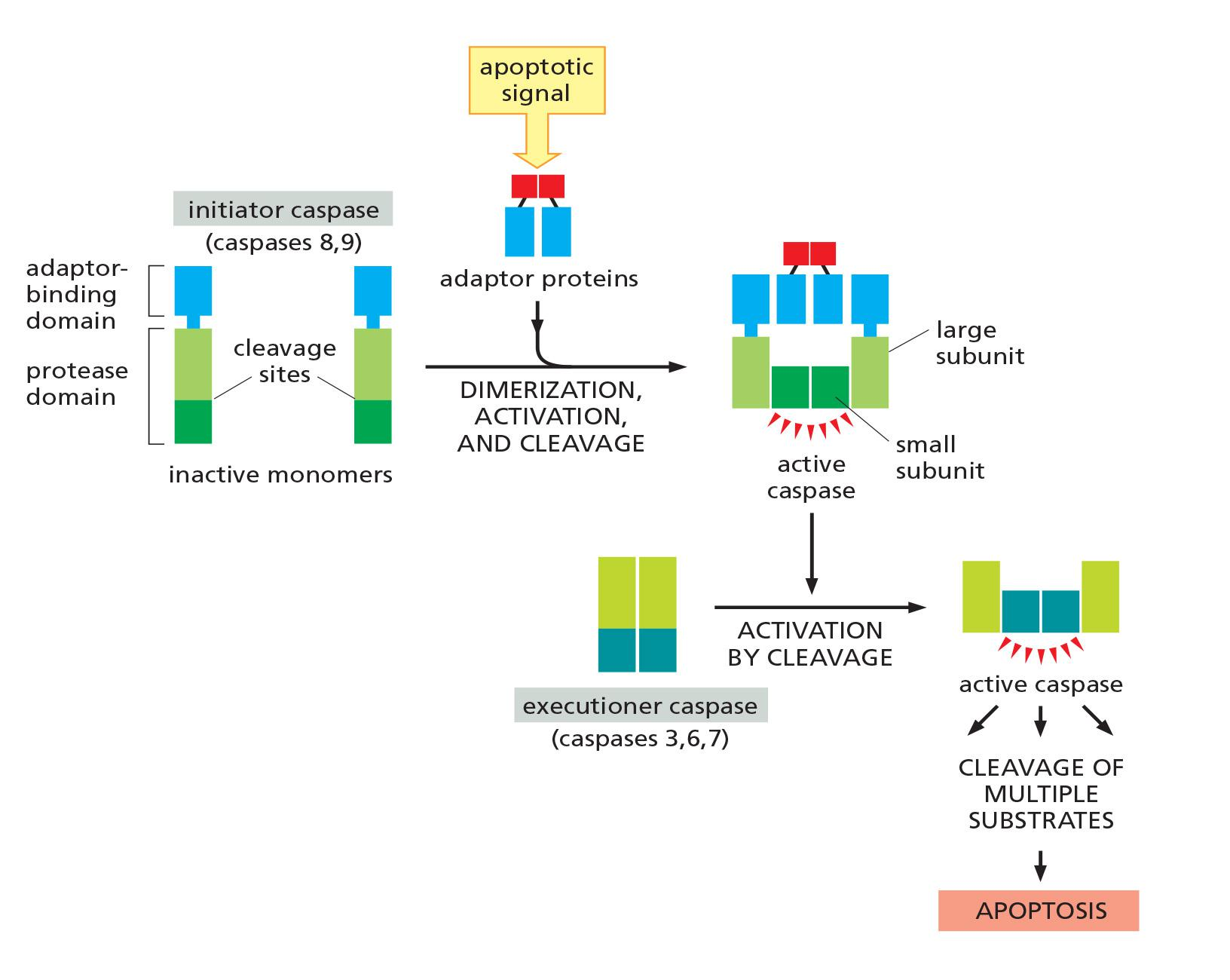
Death is dangerous: activity control is a must !!!
Activation cascade:
- Apoptotic signal triggers assembly of adaptor proteins.
- Adaptor complex triggers dimerization, cleavage, and activation of the initiator caspase.
- Activated initiator caspase activates executioner caspase by cleavage.
- Activated executioner caspase cleaves multiple substrates, resulting in cell death.
1. DNA fragmentation during apoptosis
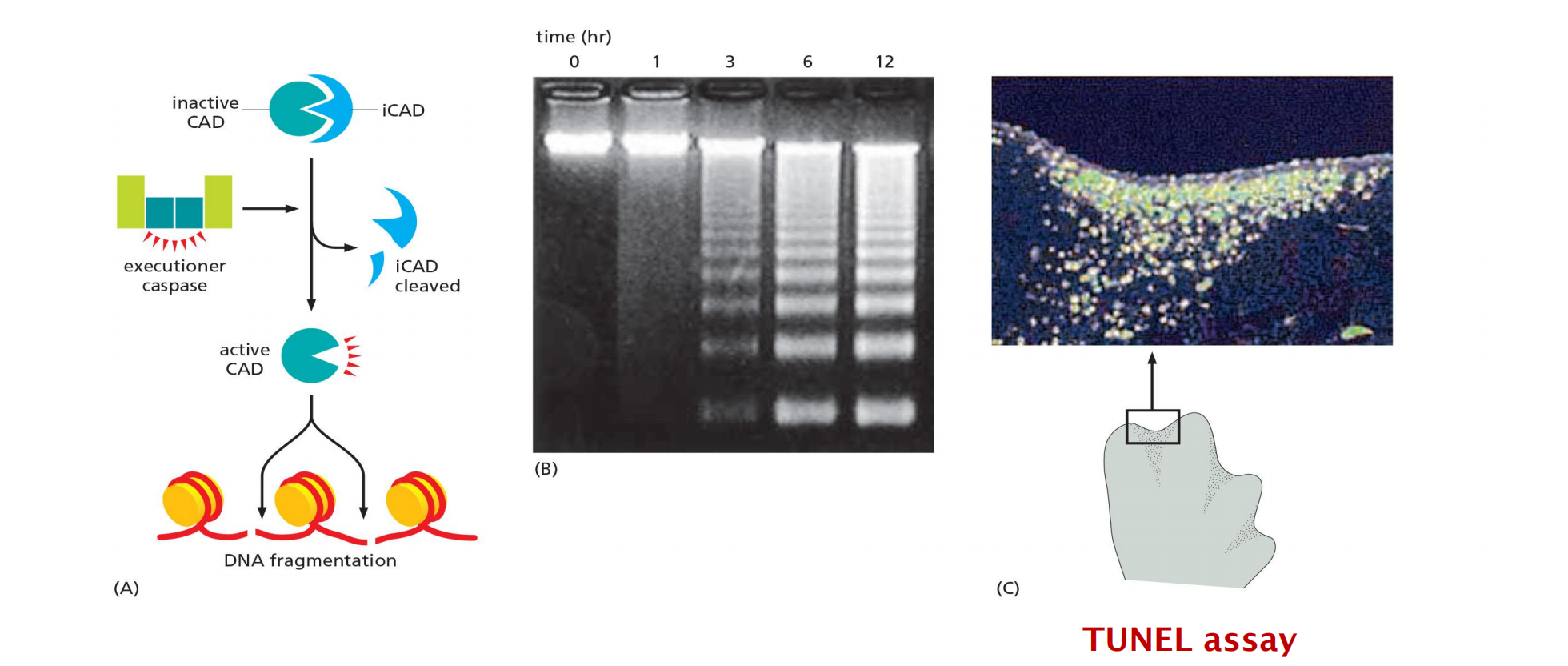
- CAD (Caspase activated deoxymonuclease): endonuclease
- iCAD: inhibitor of the endonuclease CAD
TUNEL (TdT-mediated dUTP nick end labeling) technique: the enzyme terminal deoxynucleotidyl transferase (TdT) adds chains of labeled deoxynucleotide (dUTP) to the 3ʹ-OH ends of DNA fragments.
2. The amplifying caspase cascade: the point-of-no-return
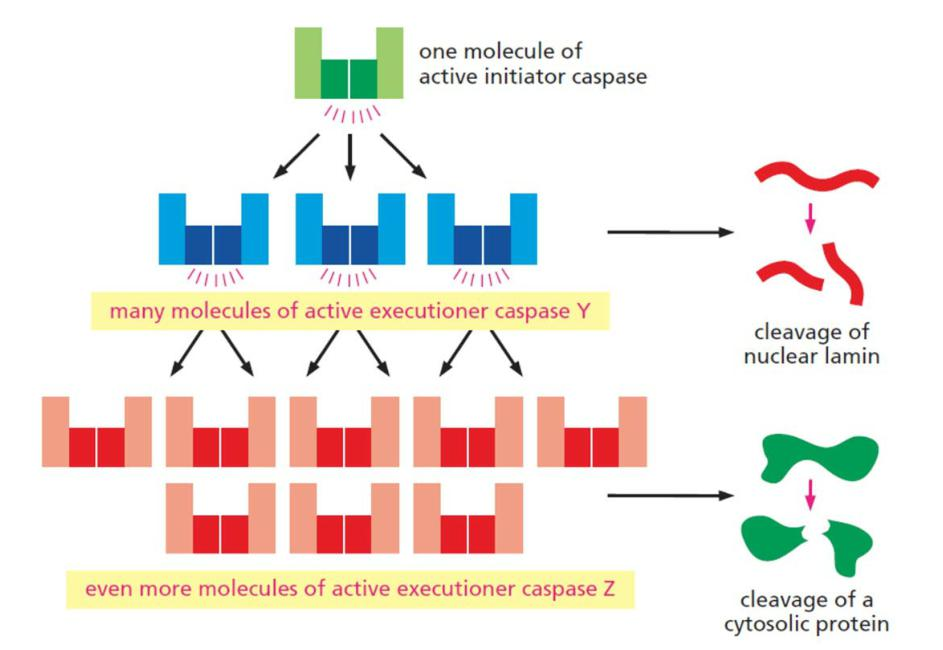
A single initiator caspase activates many molecules of executioner caspases.
3. Human caspases
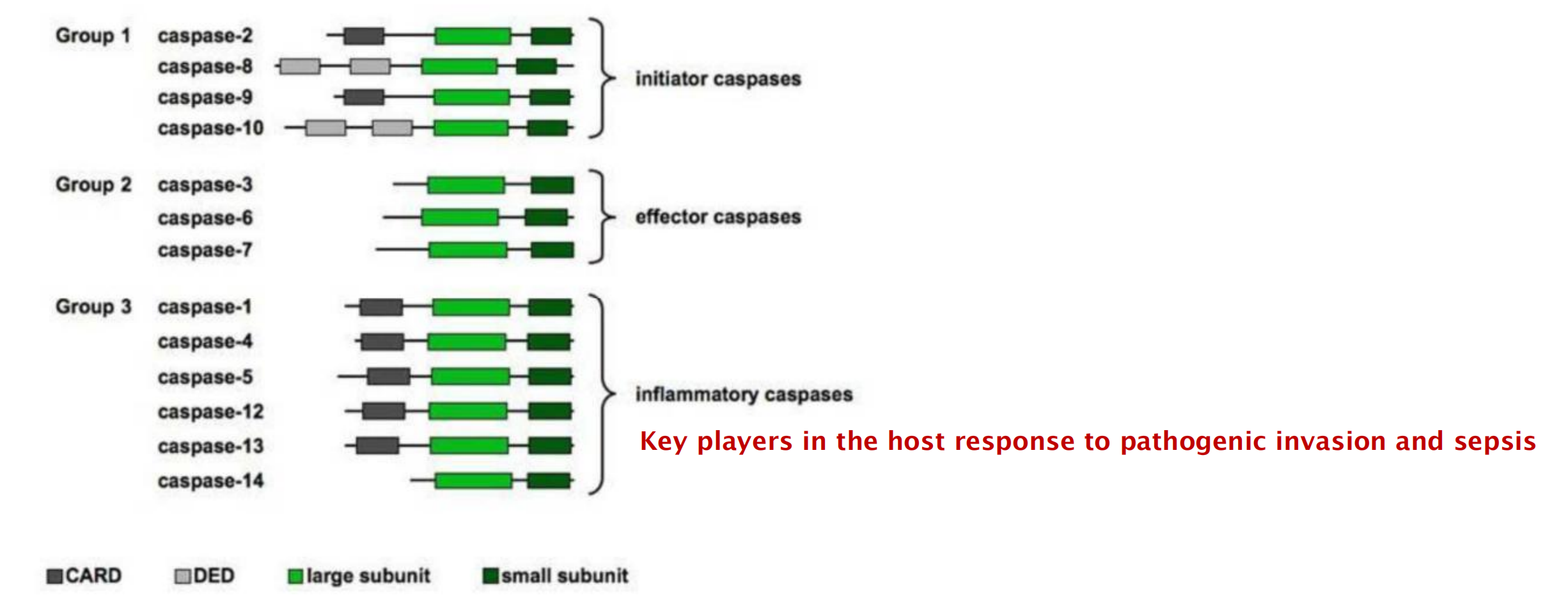
- CARD domain: caspase recruiting domain
- DED domain: death effector domain
Initiator caspases recruit adaptor proteins upon stimulations and become activated. Activated initiator caspases cleave each other and cleave effector caspases.
Extrinsic apoptosis
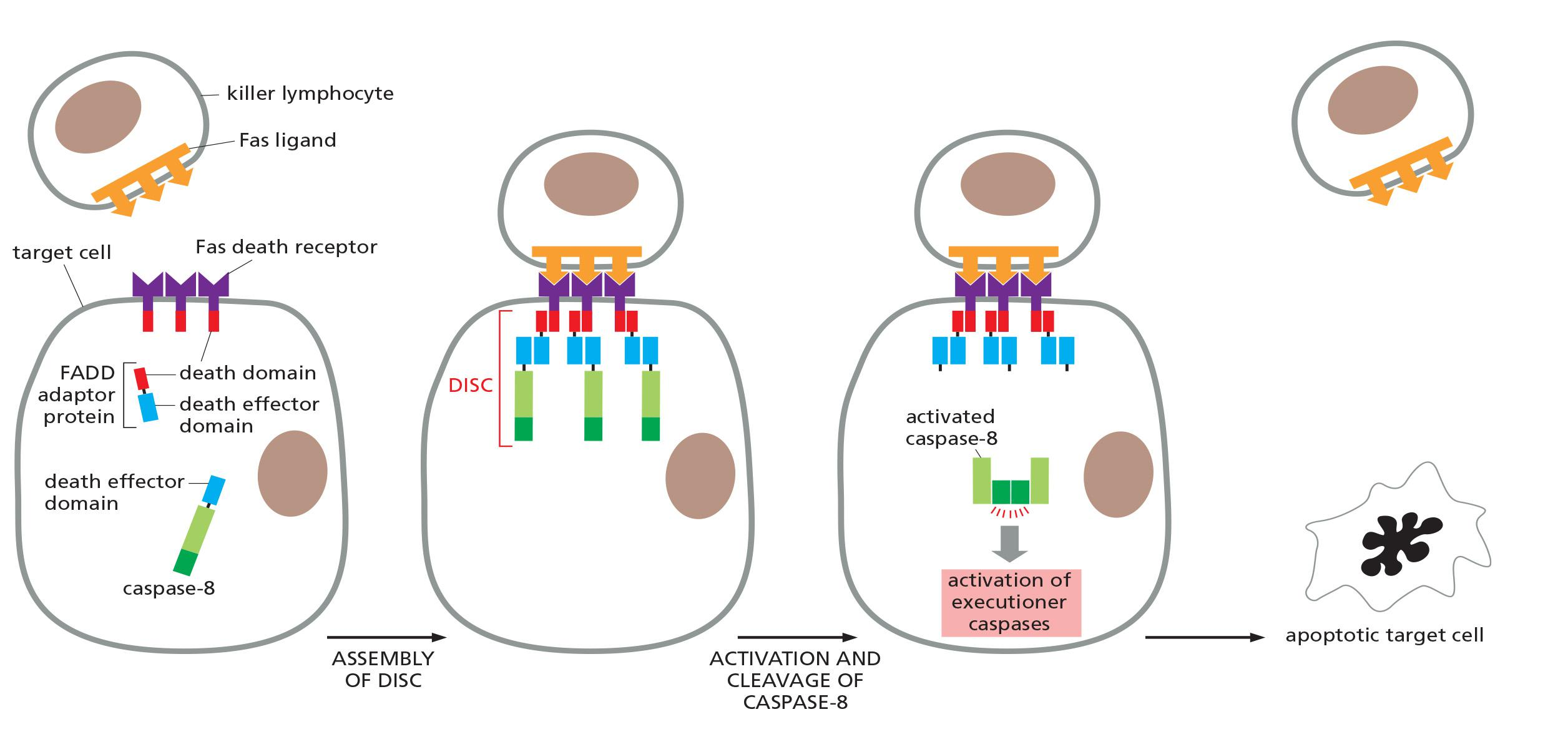
Death receptor-triggered extrinsic pathway of apoptosis:
Fas ligands (FasL): the first apoptosis signal ligand (cytotoxic T cells)
- belong to the TNF (tumor necrosis factor) family of signal proteins
- form homotrimers
- are structurally related to one another
Fas receptors (FasR):
- belongs to the TNF receptor (TNFR) family transmembrane proteins (single transmembrane domain)
- intracellular death domain (to activate the apoptosis program)
- form homotrimeric receptor complexes
Interactions:
- activated receptors recruit FADD (Fas-associating protein with a novel death domain) adaptors
- FADD adaptors recruit an initiator caspase via DED (death effector domains):
- formation of the death-inducing signaling complex (DISC)
- cleavage, activation and release of the initiator caspase
- activation of executioner caspases
Cell-Surface Death Receptors Activate the Extrinsic Pathway of Apoptosis
- Extracellular signal proteins binding to cell-surface death receptors trigger the extrinsic pathway of apoptosis
- Death receptors are transmembrane proteins containing an extracellular ligand-binding domain, a single transmembrane domain, and an intracellular death domain, which is required for the receptors to activate the apoptotic program
The receptors are homotrimers and belong to the tumor necrosis factor (TNF) receptor family, which includes a receptor for TNF itself and the Fas death receptor
- The ligands that activate the death receptors are also homotrimers; they are structurally related to one another and belong to the TNF family of signal proteins.
A well-understood example of how death receptors trigger the extrinsic pathway of apoptosis is the activation of Fas on the surface of a target cell by Fas ligand on the surface of a killer (cytotoxic) lymphocyte
- When activated by the binding of Fas ligand, the death domains on the cytosolic tails of the Fas death receptors bind intracellular adaptor proteins, which in turn bind initiator caspases (primarily caspase-8), forming a death-inducing signaling complex (DISC).
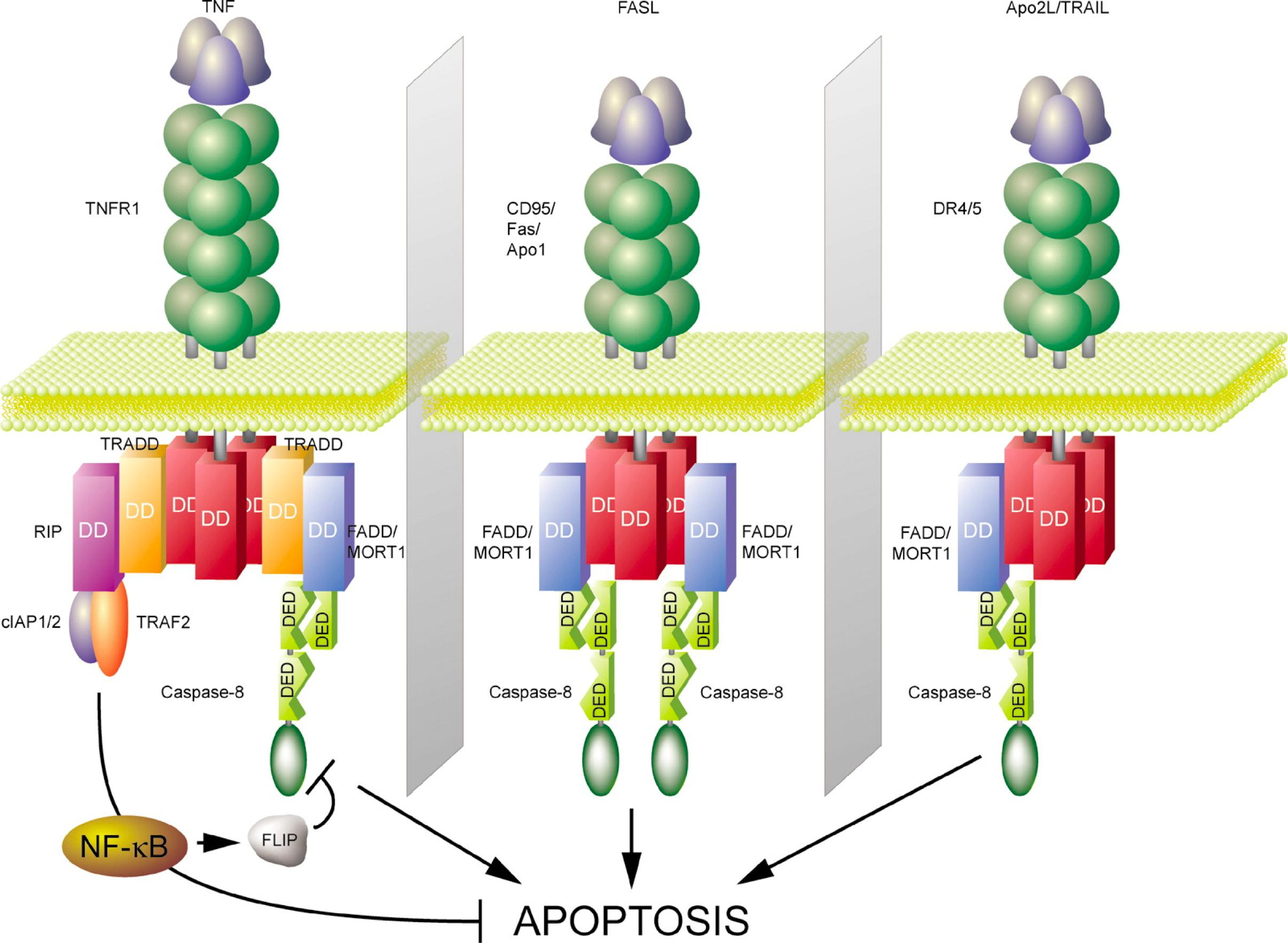
1. Inhibitor proteins in extrinsic pathway
In some cells, the extrinsic pathway recruits the intrinsic apoptotic pathway to amplify the caspase cascade and kill the cell.
Many cells produce inhibitory proteins that act to restrain the extrinsic pathway. For example, some cells produce the protein FLIP, which resembles an initiator caspase but has no protease activity because it lacks the key cysteine in its active site
The extrinsic pathway can be restrained by inhibitor proteins:
- Inhibitor proteins e.g. c-FLIP (cellular FLICE-like inhibitory protein) resembles initiator caspase 8, but has no protease activity.
- FLICE is caspase-8.
- Inhibitor proteins lack the crucial cysteine in their active site.
- Inhibitor proteins dimerize with caspase-8 in DISC but are not cleaved and activated → the signaling chain is interrupted
2. Summary: extrinsic death receptor pathways
How is the initiator caspase first activated in response to an apoptotic signal?
- The two best-understood activation mechanisms in mammalian cells are called the extrinsic pathway and the intrinsic, or mitochondrial pathway
Intrinsic apoptosis
The Intrinsic Pathway of Apoptosis Depends on Mitochondria
Cells can also activate their apoptosis program from inside the cell, often in response to stresses, such as DNA damage, or in response to developmental signals.
- In vertebrate cells, these responses are governed by the intrinsic, or mitochondrial, pathway of apoptosis, which depends on the release into the cytosol of mitochondrial proteins that normally reside in the intermembrane space of these organelles
- Some of the released proteins activate a caspase proteolytic cascade in the cytoplasm, leading to apoptosis.
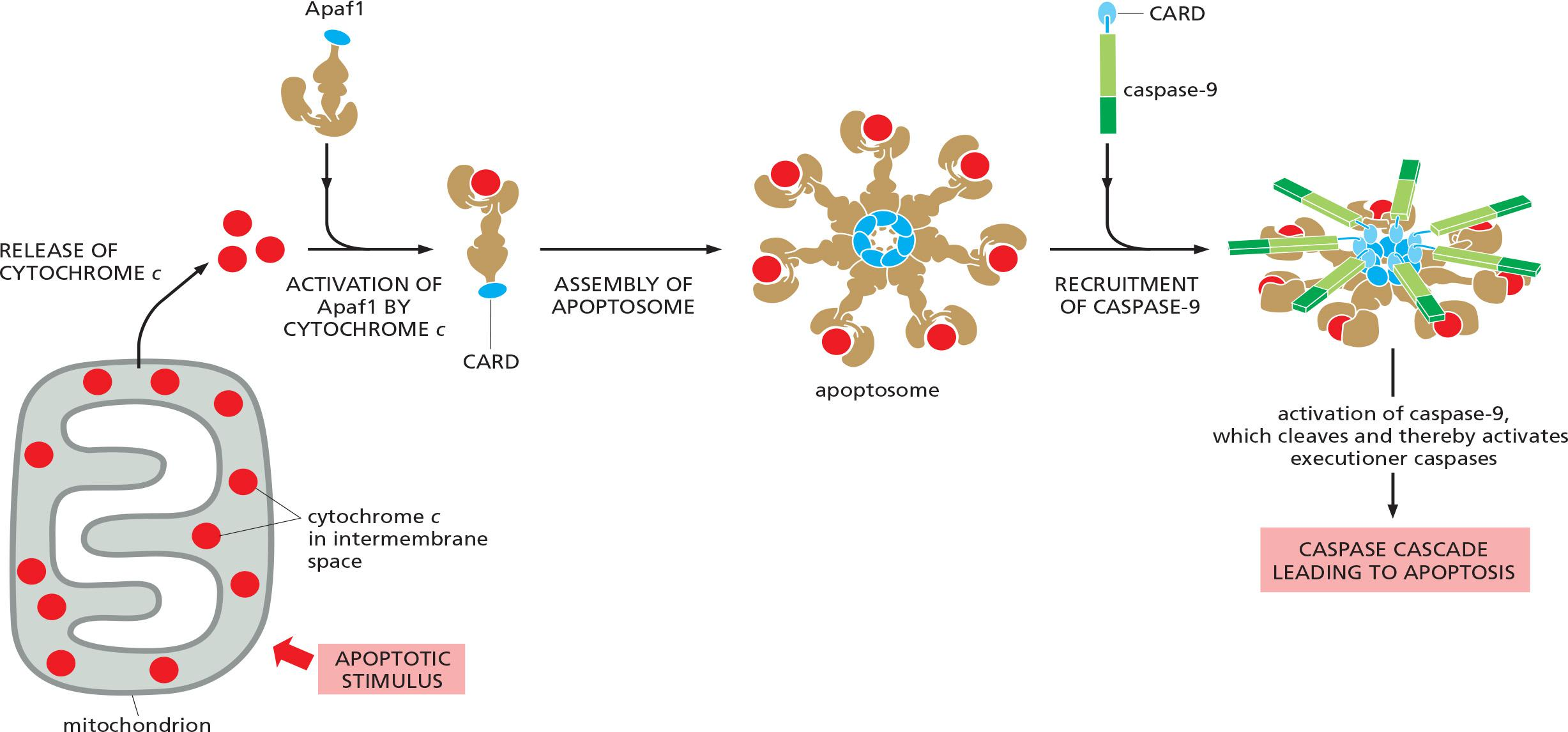
A key protein in the intrinsic pathway is cytochrome c, a water-soluble component of the mitochondrial electron-transport chain
- When released into the cytosol (Figure 18–6), it takes on a new function: it binds to an adaptor protein called Apaf1 (apoptotic protease activating factor-1), causing the Apaf1 to oligomerize into a wheel-like heptamer (七聚物) called an apoptosome (凋亡小体)
- The Apaf1 proteins in the apoptosome then recruit initiator caspase-9 proteins, which are thought to be activated by proximity in the apoptosome, just as caspase-8 is activated in the DISC.
The intrinsic apoptosis activation pathway:
- mediated by mitochondria
- triggered by protein release from the mitochondrial intermembrane space
Key players:
- cytochrome c (water soluble component of the e- transport chain)
- Apaf1 (apoptotic protease activating factor-1) containing a CARD (caspase recruitment domain) domain
- caspase-9 initiator caspase
Pathway is triggered in response to:
- DNA damage
- Hypoxia (氧不足)
- lack of nutrients
- lack of extracellular survival signals
- …
1. Apoptosome
cytochrome c binds to Apaf1, causing oligomerization
- 7 Apaf1 molecules form a wheel-like heptamer called apoptosome.
recruitment of the initiator caspase caspase-9 into the apoptosome via the CARD domains (Caspase Recruiting Domains)
- cleavage & activation of the initiator caspase, caspase-9
- activation of executioner caspases
2. Release of mitochondrial cytochrome c during apoptosis
UV-light induced DNA damage triggers release of mitochondrial cytochrome c
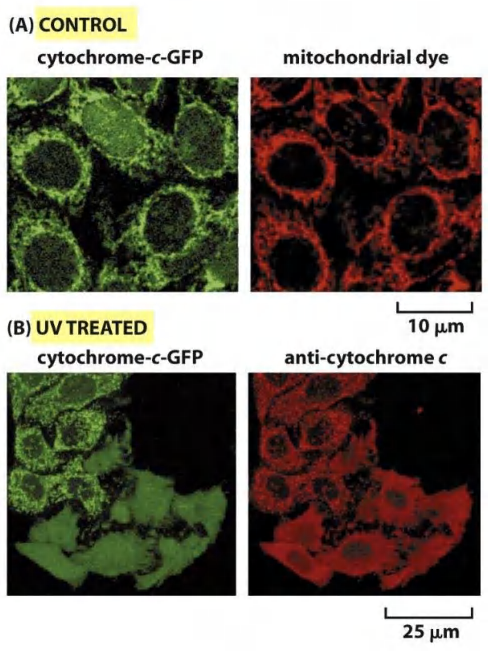
Human cancer cells expressing a mitochondrial GFP-tagged cytochrome c fusion protein (green)
Mitochondria are labeled with fluorescent dye (red) (MitoTracker™ Red)
UV-light treatment triggers release of mitochondrial proteins into the cytosol and triggers apoptosis (lower 6 cells, 5hrs after the UV treatment)
3. Bcl2 protein
Bcl2 Proteins Regulate the Intrinsic Pathway of Apoptosis
A major class of intracellular regulators of the intrinsic pathway is the Bcl2 family of proteins,
Mammalian Bcl2 family proteins regulate the intrinsic pathway of apoptosis mainly by controlling the release of cytochrome c and other intermembrane mitochondrial proteins into the cytosol
- Some Bcl2 family proteins are pro-apoptotic and promote apoptosis by enhancing the release, whereas others are anti-apoptotic and inhibit apoptosis by blocking the release.
The balance between the activities of these two functional classes of Bcl2 family proteins largely determines whether a mammalian cell lives or dies by the intrinsic pathway of apoptosis.
- As illustrated in Figure 18–8, the anti-apoptotic Bcl2 family proteins, including Bcl2 itself (the founding member of the Bcl2 family) and BclX
L, share four distinctive Bcl2 homology (BH) domains (BH1–4)
The intrinsic pathway of apoptosis is regulated by Bcl2 proteins
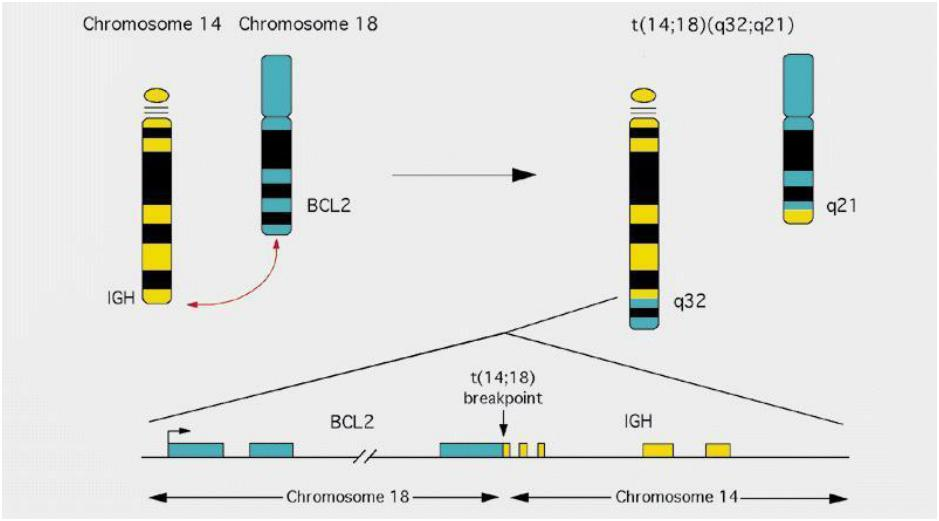
- Bcl-2 was discovered in chromosome translocation occurred in follicular lymphoma, where under control of IGH promoter, and this results in higher levels of Bcl-2, which causes cancer
The Bcl2 protein family:
- common feature: BH (Bcl2 homology) domain
- Bcl2 proteins are highly conserved
- Bcl2 proteins control the release of cytochrome c and other IMS (intermembrane space) proteins from the IMS into the cytosol
- grouped in anti-apoptotic and pro-apoptotic proteins:
- anti-apoptotic: inhibit apoptosis by blocking protein release
- pro-apoptotic: promote apoptosis by enhancing protein release
- Bcl2 proteins can form heterodimers:
- balance between inhibitory/promoting activities
- decides about life and death of the cell
Bcl2 family protein
Domain structures of members of the Bcl2 protein family
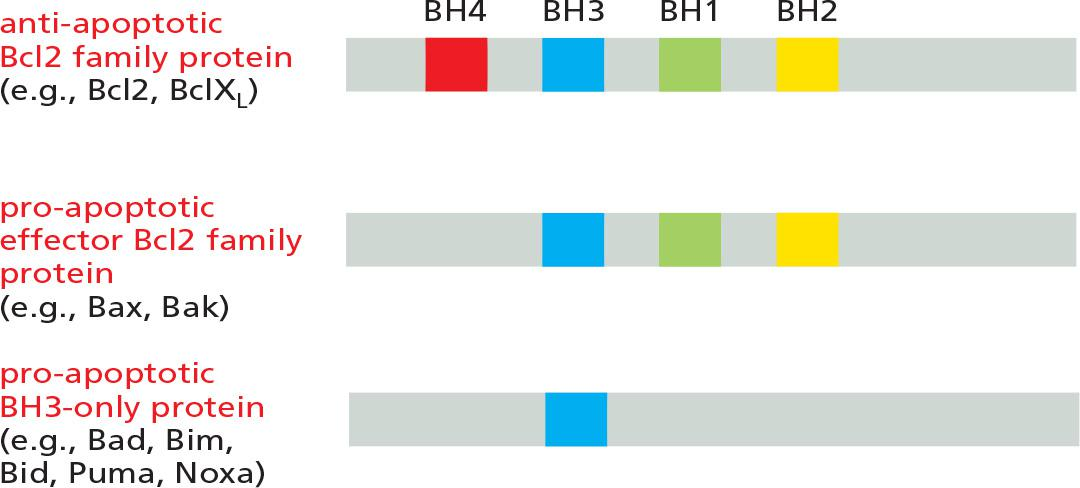
The pro-apoptotic Bcl2 family proteins consist of two subfamilies—the effector Bcl2 family proteins and the BH3-only proteins. The main effector proteins are Bax and Bak, which are structurally similar to Bcl2 but lack the BH4 domain.
- When an apoptotic stimulus triggers the intrinsic pathway, the pro-apoptotic effector Bcl2 family proteins become activated and aggregate to form oligomers in the mitochondrial outer membrane, inducing the release of cytochrome c and other intermembrane proteins by an unknown mechanism
The anti-apoptotic Bcl2 family proteins such as Bcl2 itself and **BclX
L**are also located on the cytosolic surface of the outer mitochondrial membrane
- The anti-apoptotic Bcl2 family proteins inhibit apoptosis mainly by binding to and inhibiting pro-apoptotic Bcl2 family proteins—either on the mitochondrial membrane or in the cytosol
The BH3-only proteins are the largest subclass of Bcl2 family proteins. The cell either produces or activates them in response to an apoptotic stimulus
- They are thought to promote apoptosis mainly by inhibiting anti-apoptotic Bcl2 family proteins
- Their BH3 domain binds to a long hydrophobic groove on anti-apoptotic Bcl2 family proteins, neutralizing their activity.
- This binding and inhibition enables the aggregation of Bax and Bak on the surface of mitochondria, which triggers the release of the intermembrane mitochondrial proteins that induce apoptosis
BH3-only proteins provide the crucial link between apoptotic stimuli and the intrinsic pathway of apoptosis
Similarly, in response to DNA damage that cannot be repaired, the tumor suppressor protein p53 accumulates
The BH3-only protein Bid is the link between the two pathways.
- Bid is normally inactive. However, when death receptors activate the extrinsic pathway in some cells, the initiator caspase, caspase-8, cleaves Bid, producing an active form of Bid that translocates to the outer mitochondrial membrane and inhibits anti-apoptotic Bcl2 family proteins, thereby amplifying the death signal.
4. Release of cytochrome c and IMS proteins
The pro-apoptotic effectors Bax and Bak aggregate (formation of a pore) in the outer membrane of mitochondria to release cytochrome c and IMS (Intermembrane space) proteins (e.g. endonuclease G)
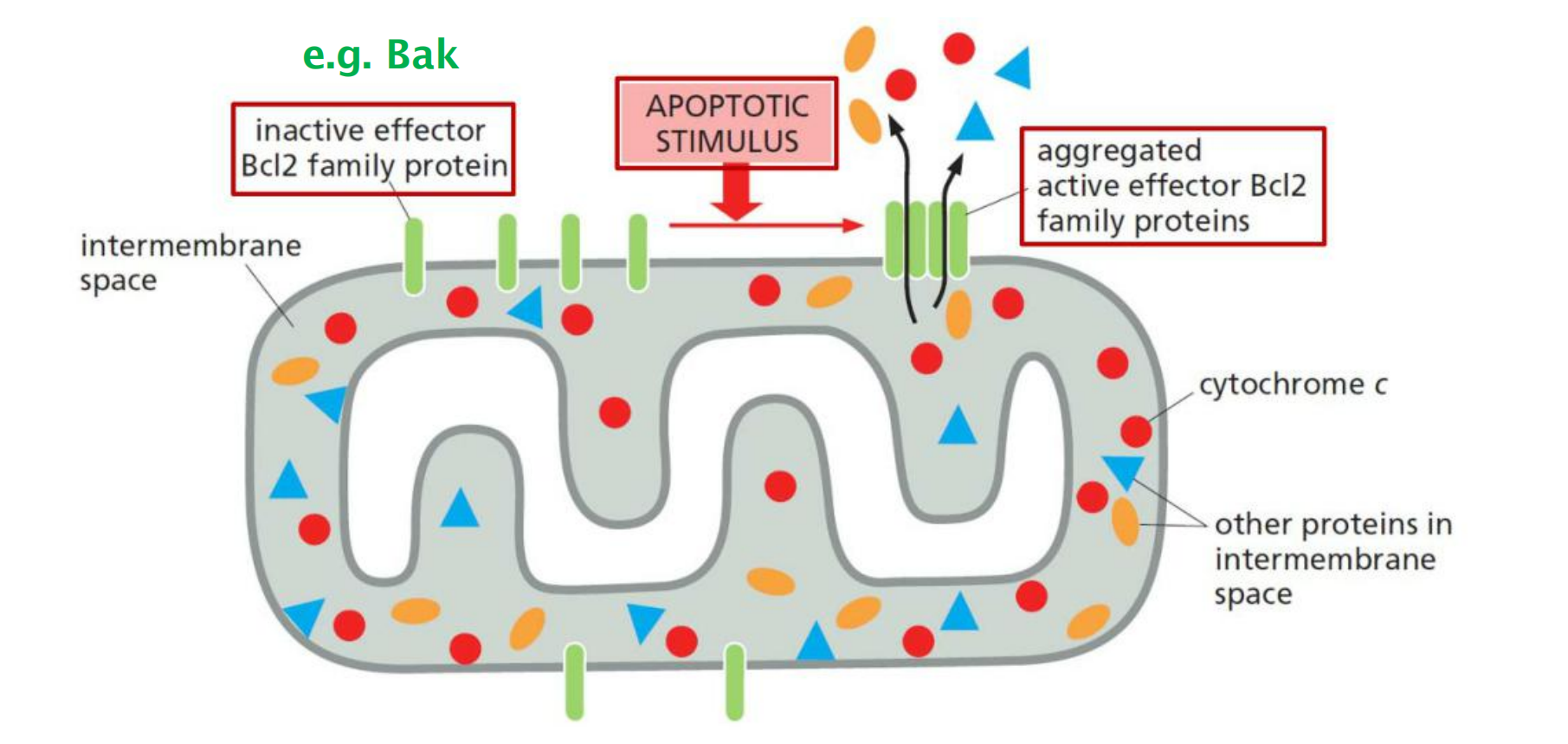
Absence of signal: Bak is mainly in the outer membrane, while Bax is cytosolic.
Upon signaling: Bax relocates to the outer membrane and interacts with Bak to trigger the efflux from the IMS peoteins
Anti-apoptotic Bcl-2 binds and inhibits Bax and Bak aggregation
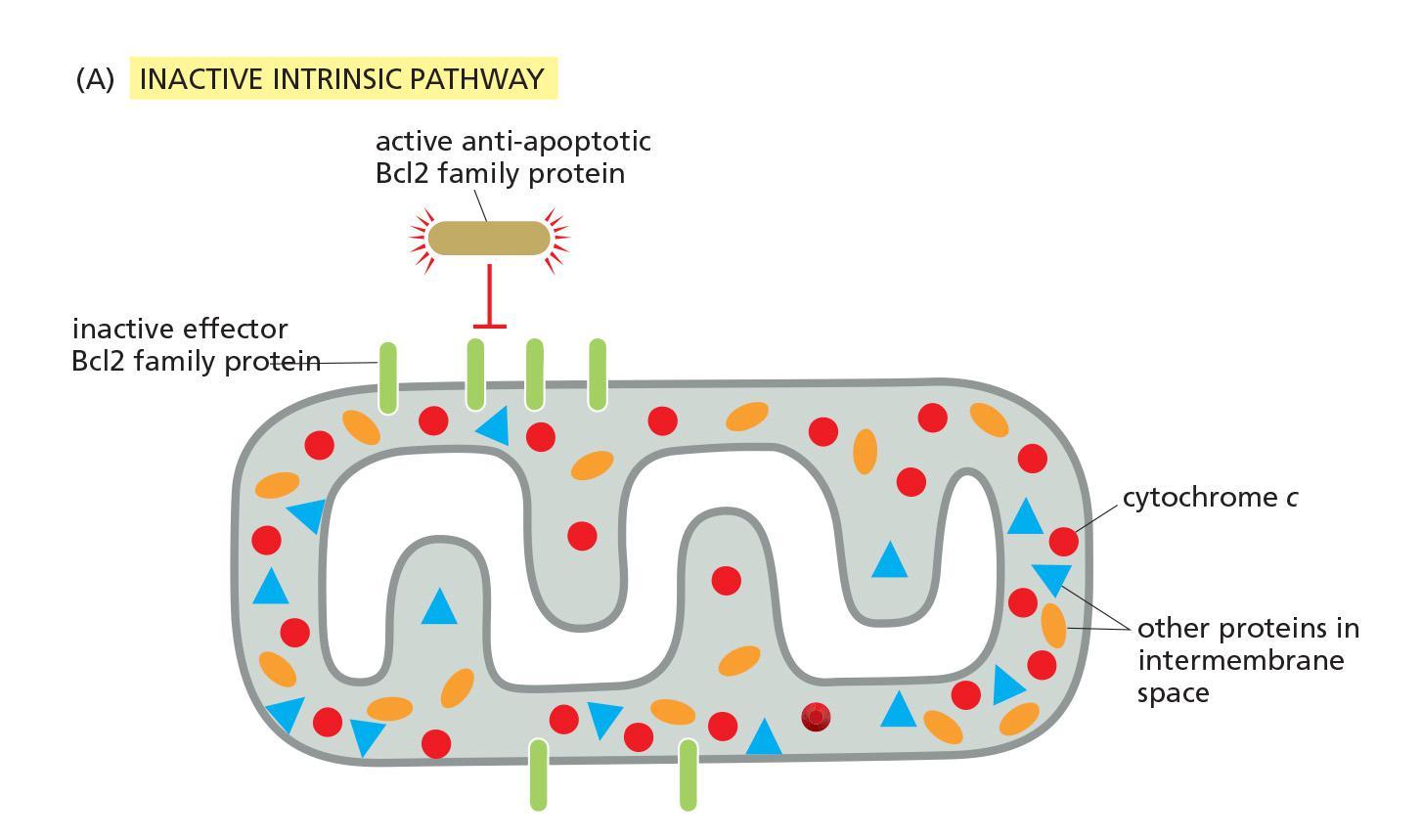
Active anti-apoptotic Bcl2 family proteins prevent aggregation of Bak-Bax at the outer mitochondrial membrane.
BH3-only proteins inhibit anti-apoptotic Bcl2 family proteins
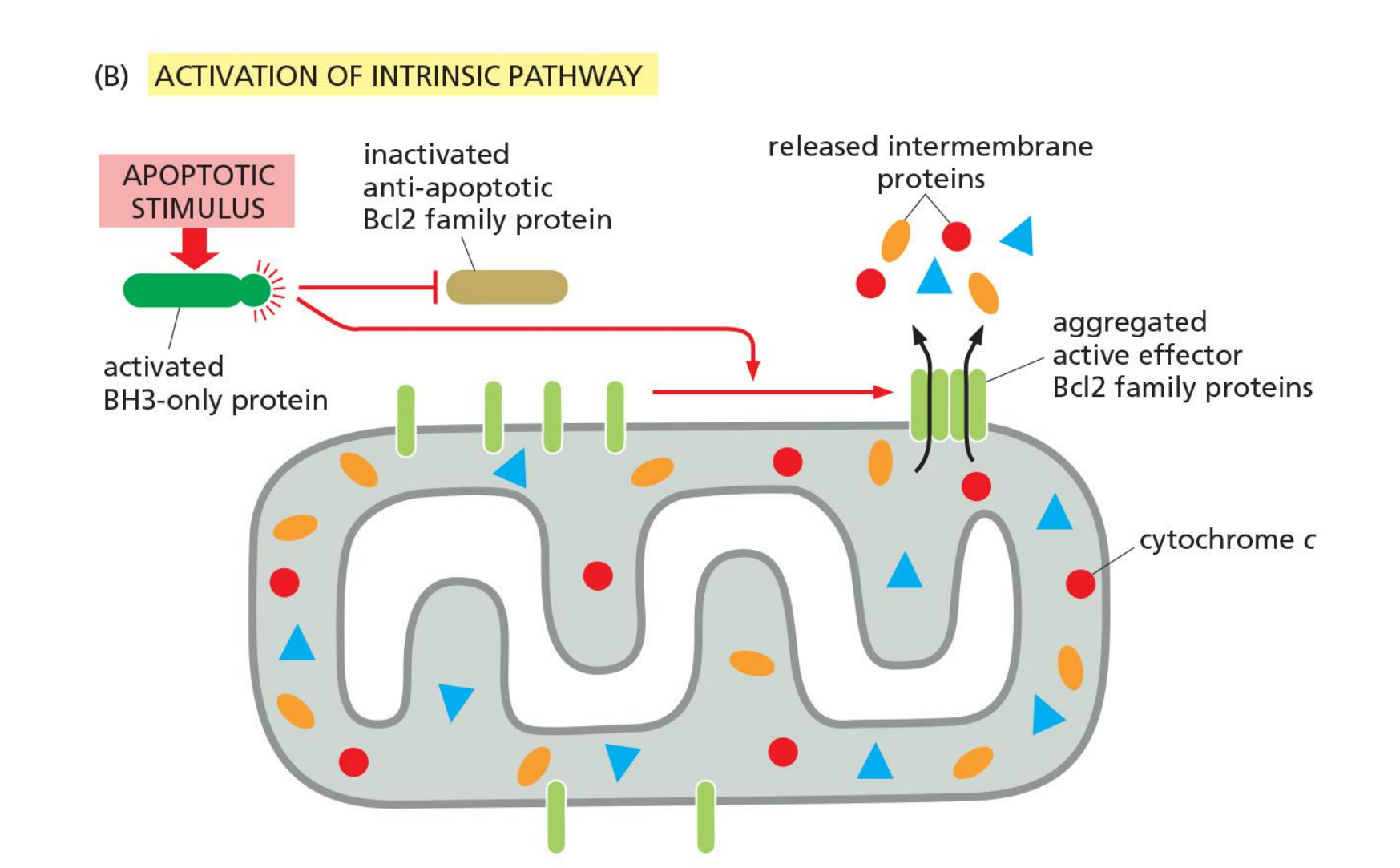
Inactivation of anti-apoptotic Bcls2 proteins by BH3-only proteins allow for Bak/Bax aggregation and thus for apoptosis.
IAPs (inhibitors of apoptosis) inhibit caspases
IAPs Help Control Caspases
- the cell employs multiple robust mechanisms to ensure that these proteases are activated only when conditions are appropriate
- One line of defense is provided by a family of proteins called inhibitors of apoptosis (IAPs)
These proteins were first identified in certain insect viruses (baculoviruses), which encode IAP proteins to prevent a host cell that is infected by the virus from killing itself by apoptosis. It is now known that most animal cells also make IAP proteins.
- All IAPs have one or more BIR (baculovirus IAP repeat) domains, which enable them to bind to and inhibit activated caspases.
- Some IAPs also polyubiquitylate caspases, marking the caspases for destruction by proteasomes. In this way, the IAPs set an inhibitory threshold that caspases must overcome to trigger apoptosis.
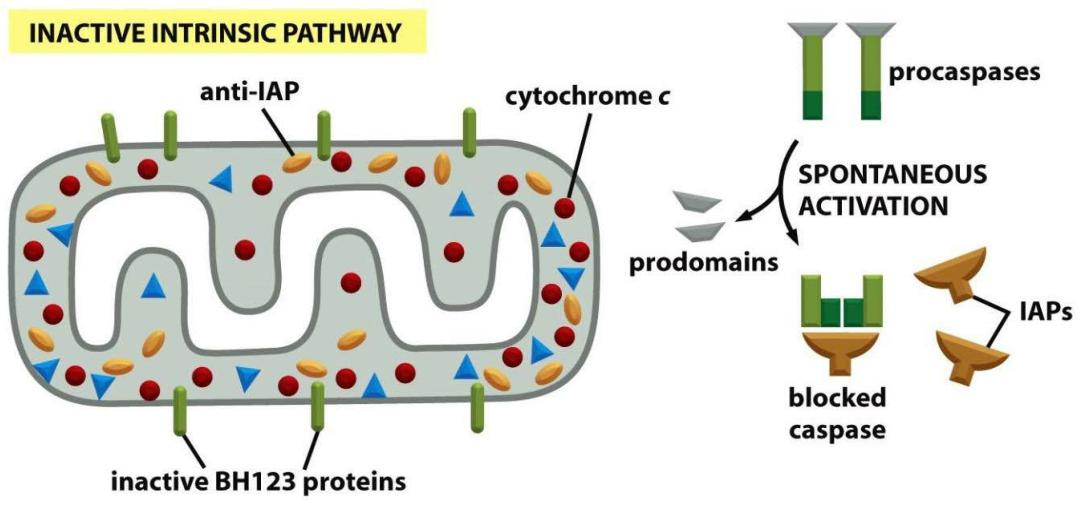
IAPs prevent the action of “accidentally” activated caspases by binding to them.
5. Summary: intrinsic mitochondrial pathways
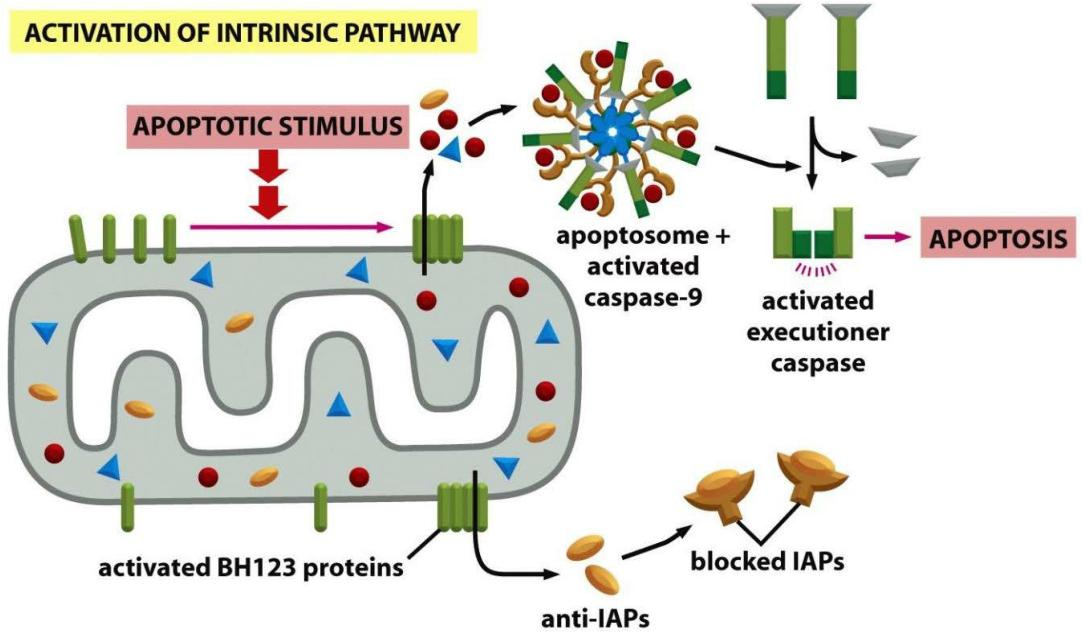
Signal: assembly of apoptosome - activation of caspases & inactivation of IAPs
Three ways for survival factors to inhibit apoptosis
Extracellular Survival Factors Inhibit Apoptosis in Various Ways
- Some extracellular signal molecules stimulate apoptosis, whereas others inhibit it
Most animal cells require continuous signaling from other cells to avoid apoptosis
Survival factors usually bind to cell-surface receptors, which activate intracellular signaling pathways that suppress the apoptotic program
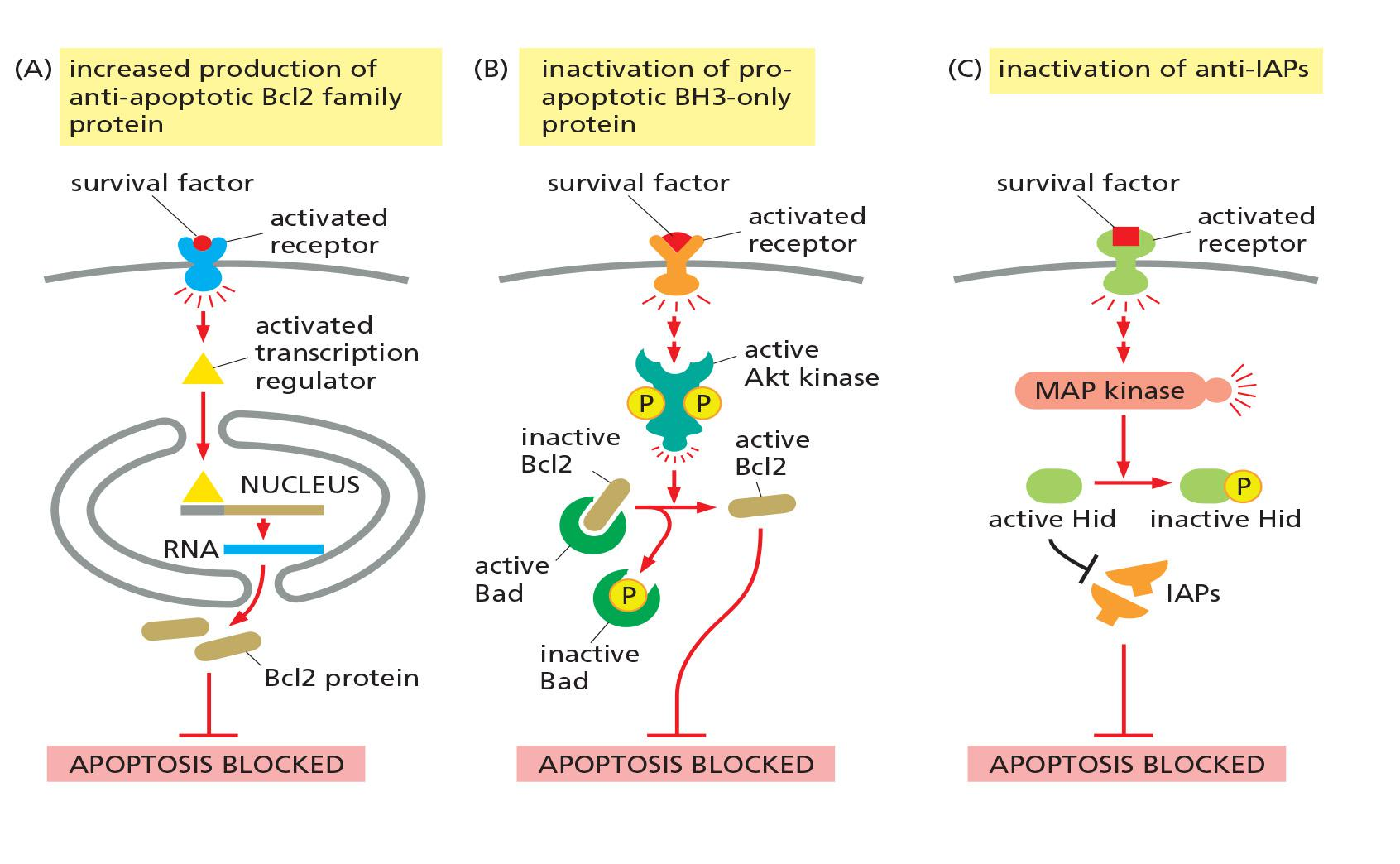
There are 3 ways:
- Increased production of anti-apoptotic proteins
- Inactivation of pro-apoptotic protein - BH3-only proteins
- Inactivation of anti-IAPs
1. The role of survival factors and cell death in adjusting the number of cells
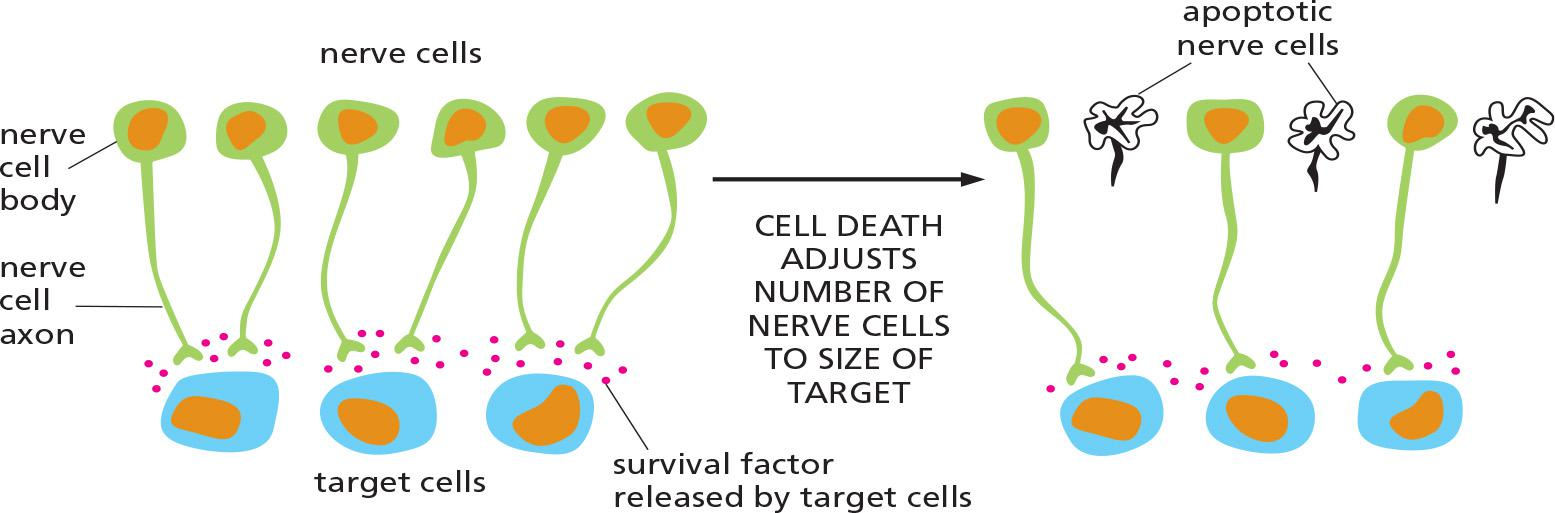
Apoptosis is indispensable for successful development, and maintenance of homeostasis within multicellular organisms.
More nerve cells are produced than can be supported by the limited amount of survival factors released by the target cells.
This strategy of overproduction followed by culling (选择;大批物品中剔出劣质货) helps ensure that all target cells are contacted by nerve cells.
Phagocytes remove the apoptotic cells
Phagocytes Remove the Apoptotic Cell
This engulfment process depends on chemical changes on the surface of the apoptotic cell, which displays signals that recruit phagocytic cells
- An especially important change occurs in the distribution of the negatively charged phospholipid phosphatidylserine (PS) on the cell surface.
This phospholipid is normally located exclusively in the inner leaflet of the lipid bilayer of the plasma membrane (see Figure 10–15), but it flips to the outer leaflet in apoptotic cells
The external exposure of phosphatidylserine is likely to depend on caspase cleavage of some protein involved in phospholipid distribution in the membrane.
Healthy cells express signal proteins on their surface that interact with inhibitory receptors on macrophages that block phagocytosis
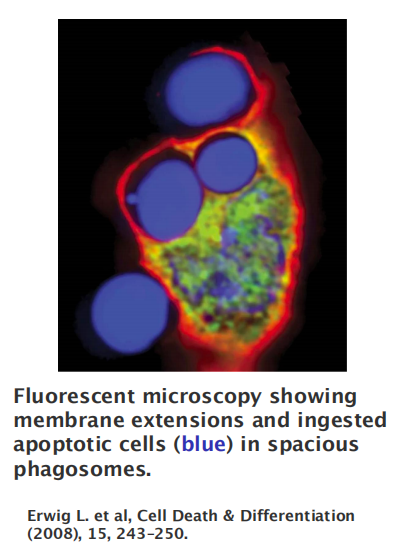
Apoptotic cells are efficiently eaten—or phagocytosed —by neighboring cells
Macrophages and dendritic cell, and other cells known to have phagocytic capacity (e.g., hepatocytes, endothelial cells, epithelial cells)
Phagocytosis is an extremely complex process and no single model can account for the diverse structures and outcomes associated with particle ingestion.
In principle, it includes:
- receptor-mediated particle recognition (negatively charged phospholipid phosphatidylserine (PS) on the cell surface)
- actin polymerization at the site of ingestion and the particle internalization
- the phagosome trafficking in association with microtubules
- The phagosome maturation by a series of fusion and fission events with components of the endocytic pathway culminating in the mature phagolysosome.
Deregulation of apoptosis causes disease
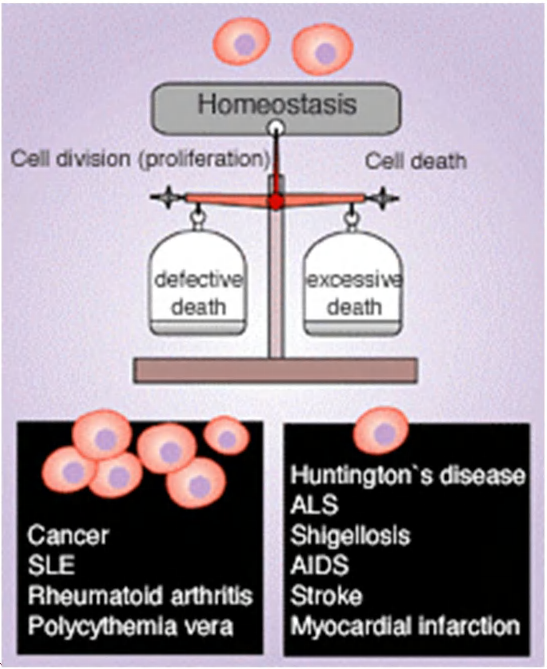
High activity of apoptosis:
- neurodegenerative disease
- myocardial infarction (心肌梗死)
- radiation injury (辐射损伤)
Low activity of apoptosis:
- cancer
- autoimmune disease
1. Tumor suppressor: P53 (Lecture 1)
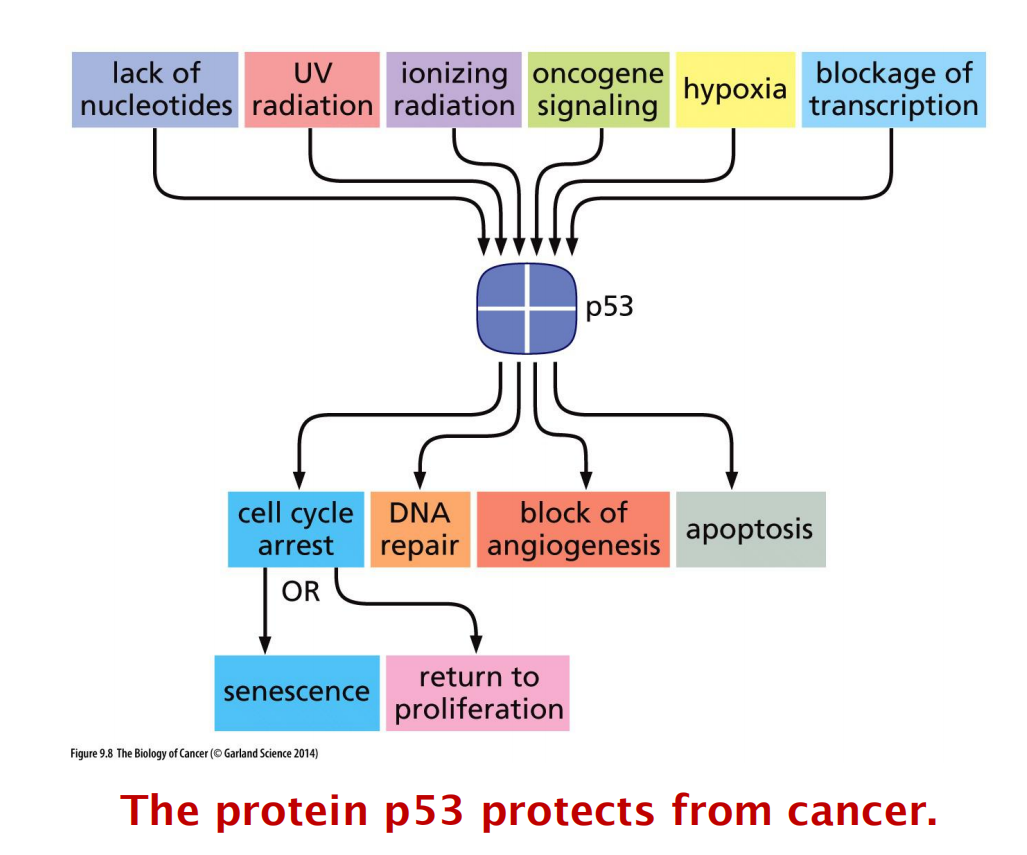
How can one single protein protect cells from cancer progression?
2. Bcl2 inhibitor: ABT-737
三、Study apoptosis
- Annexin V staining
- Cell cycle distribution
- DNA fragmentation
- TUNEL assay
- Western blot for apoptosis markers
- Cytochrome c translocation ( Caspase 3, PARP, etc)
Annexin V staining ( early apoptosis)
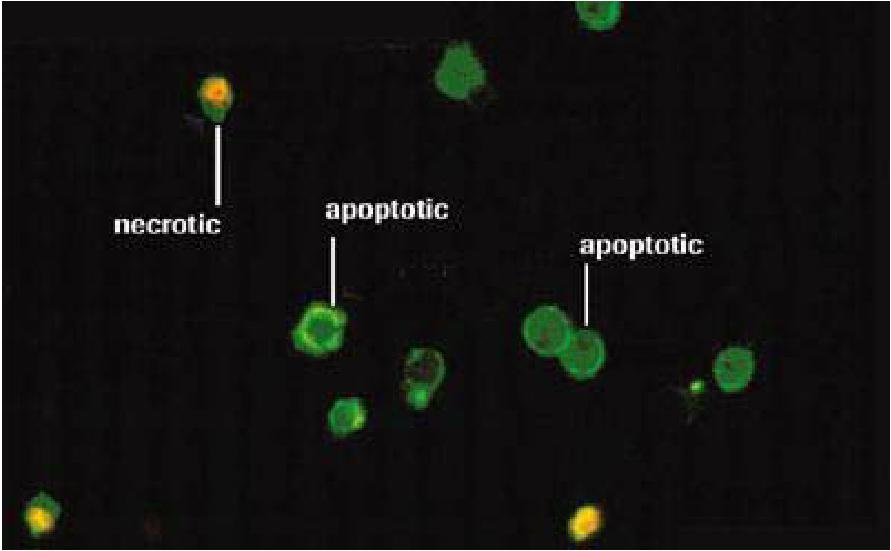
Phosphatidylserine (PS) is flipped to outer leaflet of plasma membrane and can be detected by annexin V (protein)-conjugate fluorescent dye to indicate early apoptosis
Cell cycle distribution

Sub-G1 percentage is an indicator for DNA fragmentation due to apoptosis.
DNA fragmentation
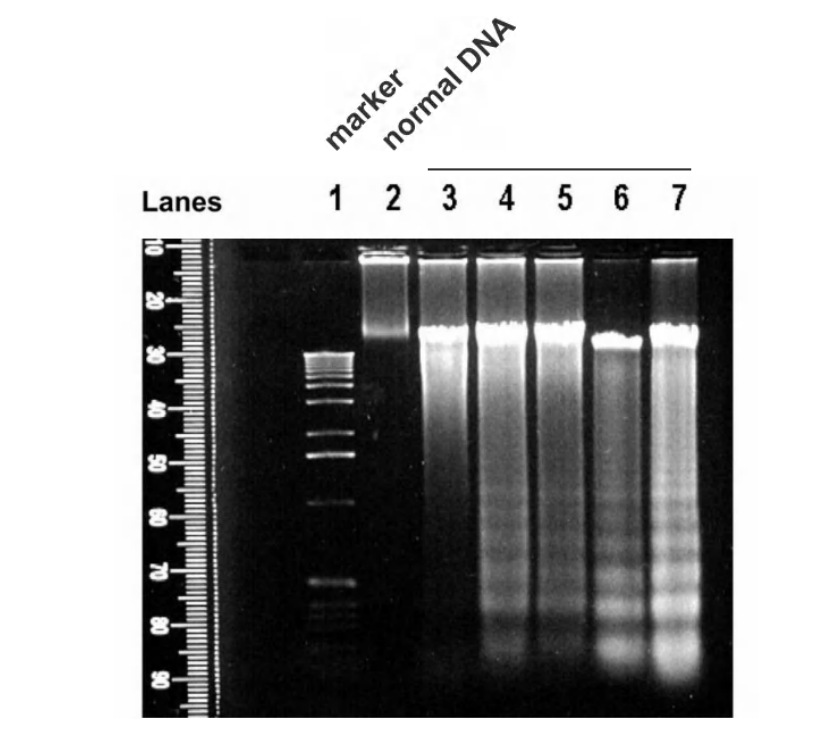
TUNEL assay (late apoptosis)
Terminal dUTP Nick End Labelling (TUNEL) staining is to detect DNA fragmentation (late apoptosis)
It is another method to detect DNA fragmentation
Terminal deoxynucleotidyl transferase catalyzes dUTPaddition on fragmented DNA, dUTP is subsequently labeled with fluorescence dye
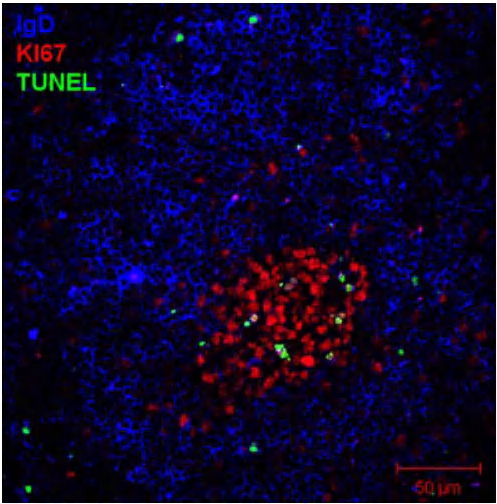
Spleen from NP-KLH immunized mice stained with:
anti-IgD (blue) → B cell follicle
anti-KI67 (red) → germinal center B cells
TUNEL (green) → counter-selected B cells that dye in the germinal center by apoptosis (positive for TUNEL stain)
Apoptosis marker for Western blotting
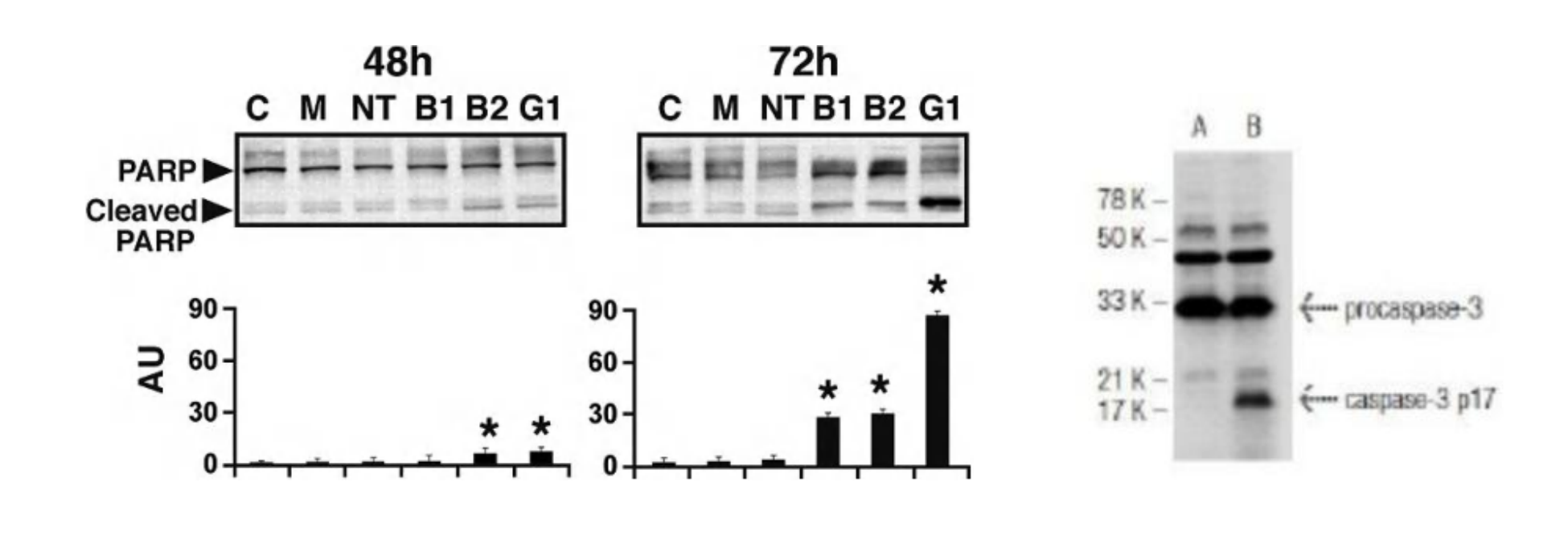
Commonly used proteins:
- cleaved PARP
- cleaved caspase-3