L11 DNA repair and recombination
一、Homologous Recombination
Essential in meiosis for generating diversity and for chromosome segregation, and in mitosis to repair DNA damage and stalled replication forks.
Homologous recombination can occur at any point along the lengths of two homologous DNAs.
Types of Recombination
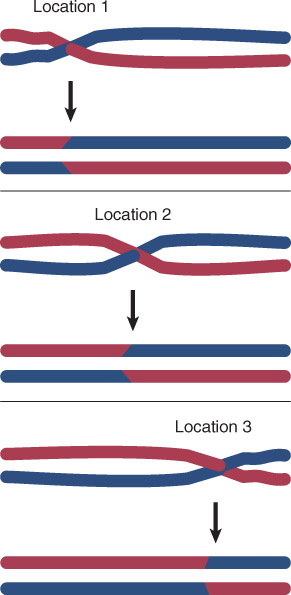
Site-specific recombination involves specific DNA sequences.
- Site-specific recombination occurs between two specific sequences (identified by boxes). The other sequences in the two recombining DNAs are not homologous.
Somatic recombination – Recombination that occurs in nongerm cells (i.e., it does not occur during meiosis); commonly in the immune system, may occur in plant vegetative tissues.
Occurance Location
Homologous recombination occues between synapsed chromosomes in meiosis(减数分裂)
Chromosomes must synapse (pair) in order for chiasmata(染色体交叉) to form where crossing over occurs
Stages of meiosis are correlated with the molecular events that occur to DNA.
Recombination involves pairing between complementary strands of the two parental DNAs.
sister chromatid – Each of two identical copies of a replicated chromosome; this term is used as long as the two copies remain linked at the centromere.
- Sister chromatids separate during anaphase in mitosis or anaphase II in meiosis.
bivalent – The structure containing all four chromatids (two representing each homolog) at the start of meiosis
synaptonemal complex – The protein structure that forms between synapsed homologous chromosomes that is believed to be necessary for recombination to occur
joint molecule – A pair of DNA duplexes that are connected together through a reciprocal exchange of genetic material.
1. Recombining Chromosomes Are Connected by The Synaptonemal Complex
During the early part of meiosis, homologous chromosomes are paired in the synaptonemal complex.
The mass of chromatin of each homolog is separated from the other by a proteinaceous complex.
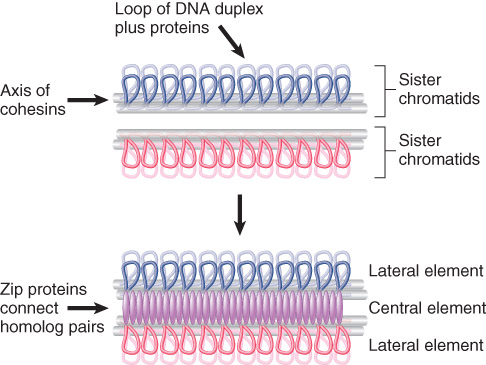
Each pair of sister chromatids has an axis made of cohesins.
Recombination Initiation
Doublr-Strand breaks initiate recombination
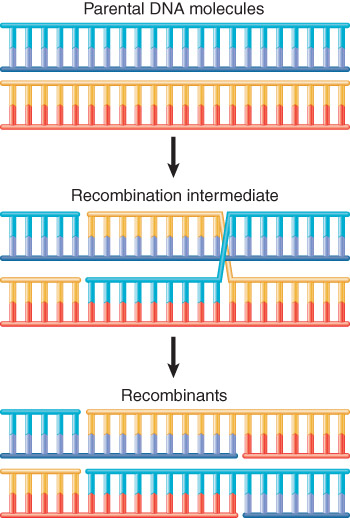
The double-strand break repair (DSBR) model of recombination is initiated by making a double-strand break in one (recipient) DNA duplex.
In 5’ end resection (5’末端切除), exonuclease action generates 3′ single-stranded ends that invade the other (donor) duplex.
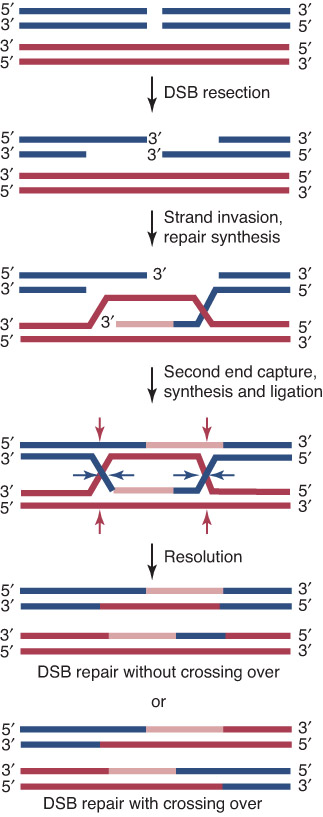
When a single strand from one duplex displaces its counterpart in the other duplex (single strand invasion), it creates a D-loop
The exchange generates a stretch of heteroduplex DNA (异源双链DNA) consisting of one strand from each parent
Double-strand break repair model of homologous recombination.
New DNA synthesis replaces any material that has been degraded
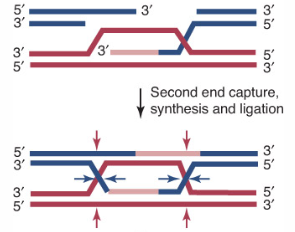
This generates a recombinant joint molecule in which the two DNA duplexes are connected by heteroduplex DNA and two Holliday junctions (霍利迪路口).
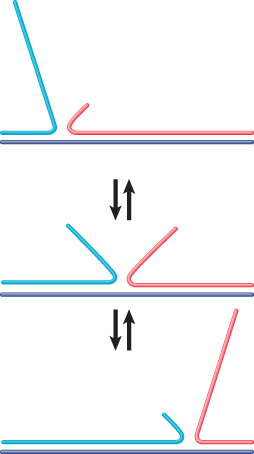
Branch migration – The ability of a DNA strand partially paired with its complement in a duplex to extend its pairing by displacing the resident strand with which it is homologous.
Branch migration can occur in either direction when an unpaired single strand displaces a paired strand.
Whether recombinants are formed depends on whether the strands involved in the original exchange or the other pair of strands are nicked during resolution.
Holliday Junctions Must Be Resolved
The Ruv complex acts on recombinant junctions.
RuvA recognizes the structure of the junction and RuvBis a helicase that catalyzes branch migration.
RuvC cleaves junctions to generate recombination intermediates.
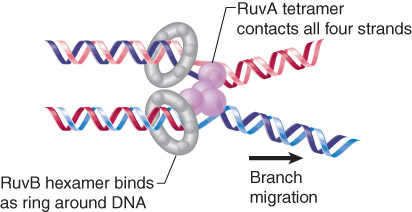
RuvAB is an asymmetric complex that promotes branch migration of a Holliday junction.
Topoisomerases
Topoisomerases relax or introduce supercoils in DNA
1. Type I
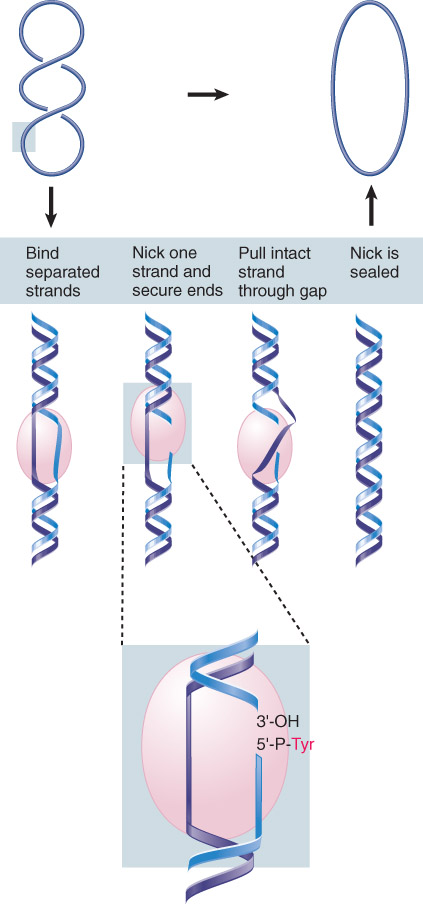
Type I topoisomerases function by forming a covalent bond to one of the broken ends, moving one strand around the other and then transferring the bound end to the other broken end. Bonds are conserved; as a result, no input of energy is required.
Type I topoisomerases recognize partially unwound segments of DNA and pass one strand through a break made in the other
2. Type II
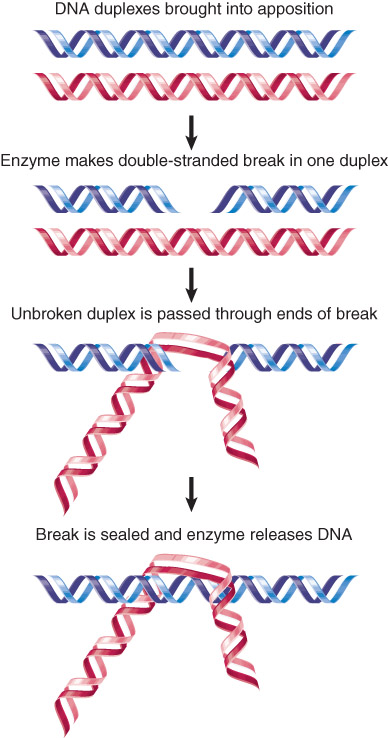
Type II topoisomerases also form covalent bonds to the broken ends, and pass a duplex DNA region through the double-strand break. ATP is required to complete the reaction and reseal the break.
Type II topoisomerases can pass a duplex DNA through a double-strand break in another duplex.
3. Gyrase And Reverse Gyrase
gyrase – Enzyme that introduces negative supercoils into DNA.
reverse gyrase – Enzyme that introduces positive supercoils into DNA
4. Site-Specific Recombination Resembles Toposomerase Activity
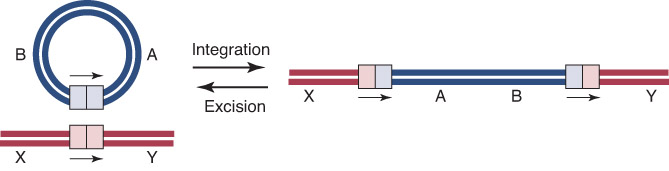
Site-specific recombination involves a reaction between specific sites that are not necessarily homologous.
recombinase – Enzyme that catalyzes site-specific recombination.
Phage lambda integrates into the bacterial chromosome by recombination between a site on the phage and the attachment (att) site on the E. coli chromosome.
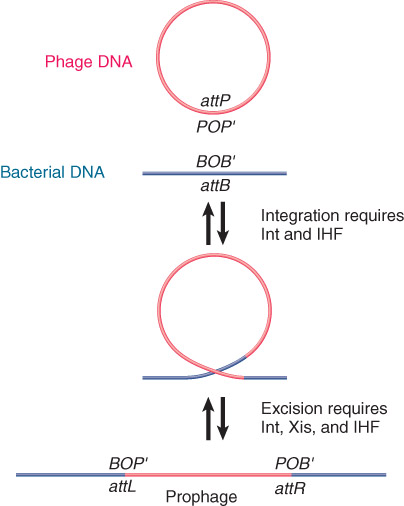
Circular phage DNA is converted to an integrated prophage by a reciprocal recombination between attP and attB.
二、Mutations
Definitions
Mutation = heritable change in sequence of DNA, particulariy a substitution, addition or deletion of one or more bases.
Mutagen = a physical or chemical agent that causes mutations or increases their occurrence.
Point Mutations Can Be Permanently Incorporated Via DNA Replication
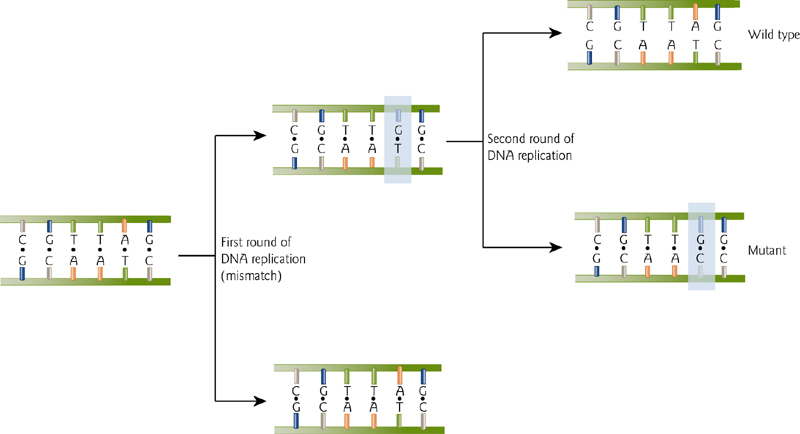
Uncorrected mutations will be retained.
Reversion of Mutations
Back mutations or “reversions” are the reverse process of mutation. This can occur by a change at the same site as the original mutation, or by a change at a different site.
Changes at different sites that ‘correct’ the mutation are called “second-site” or “suppressor” mutations, and can occur within the same gene or even in an entirely different gene.
An example might be a reversion of a frameshift mutation:
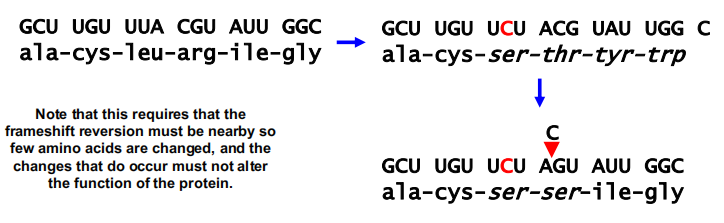
Note that this requires that the frameshift reversion must be nearby so few amino acids are changed, and the changes that do occur must not alter the function of the protein
Nucleotide Substitutions
Point mutations change one nucleotide for another.
(1) “transition” - purine replaces a purine (A ↔ G) or a pyrimidine replaces a pyrimidine, (C ↔ T). Caused by nitrous acid, base mis-pairing, or mutagenic base analogs such as 5-bromo-2-deoxyuridine (BrdU).
(2) “transversion” a purine replaces a pyrimidine or a pyrimidine replaces a purine (C/T ↔ A/G).
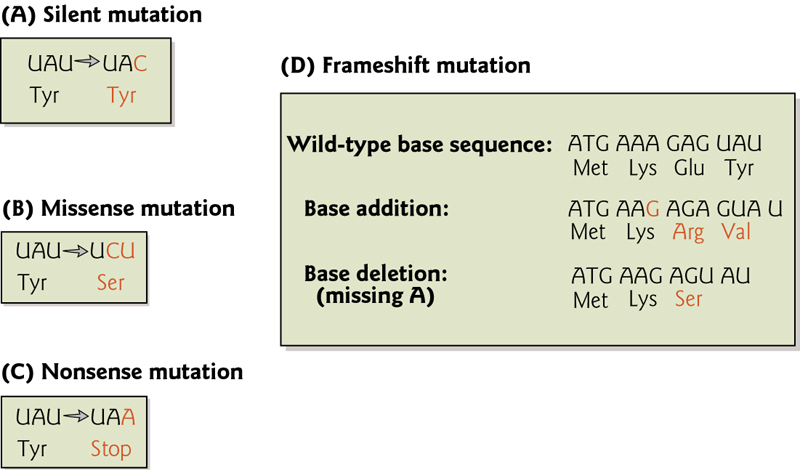
Types of DNA Damage
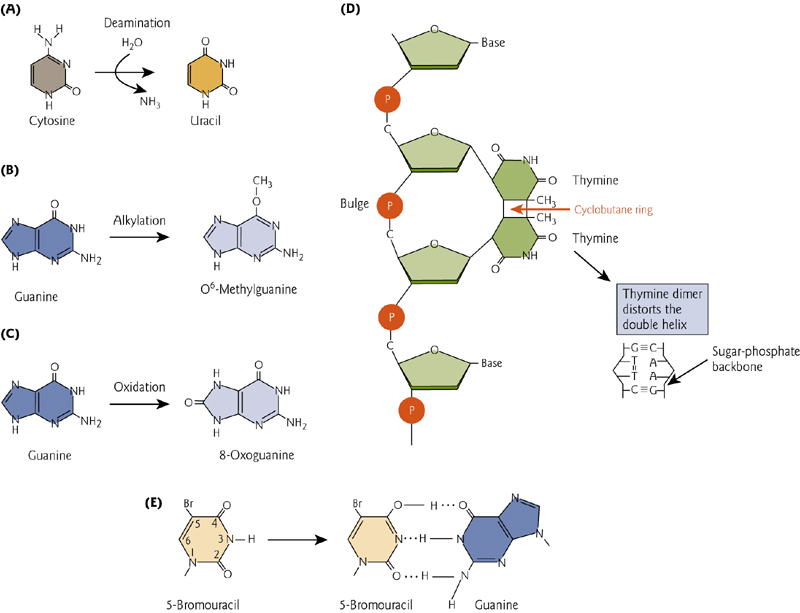
三、DNA Repair
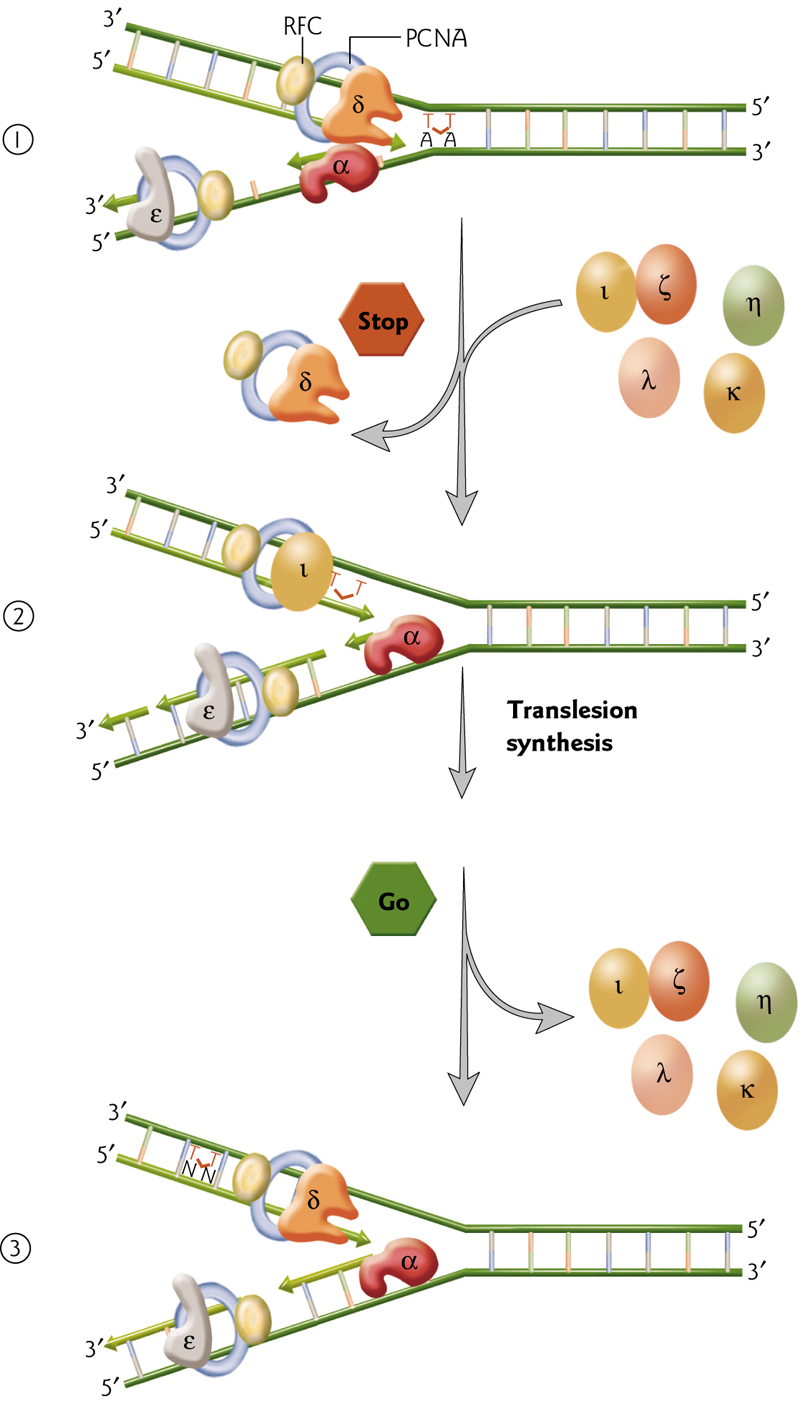
Translation DNA Repair
Normal, high fidelity DNA polymerases cannot bypass DNA lesions that distort the structure. This can be done by low fidelity polymerases that temporarily replace the high-fi pols, copy past, and then are replaced again.
- Replication machinery pauses at a thymine dimer, and polymerases are temporarily substituted.
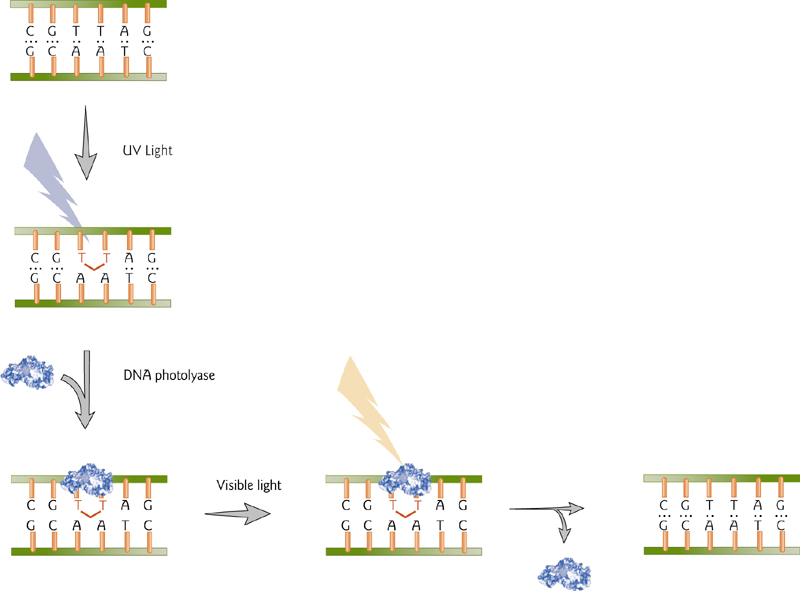
DNA Repair by Photoreactivation
A direct repair mechanism is the enzyme-mediated reversal of pyrimidine dimers that are formed by UV light. This is accomplished by DNA photolyase, an enzyme that utilizes visible light to cleave the dimer.
DNA Repair by Methyl Group Removal
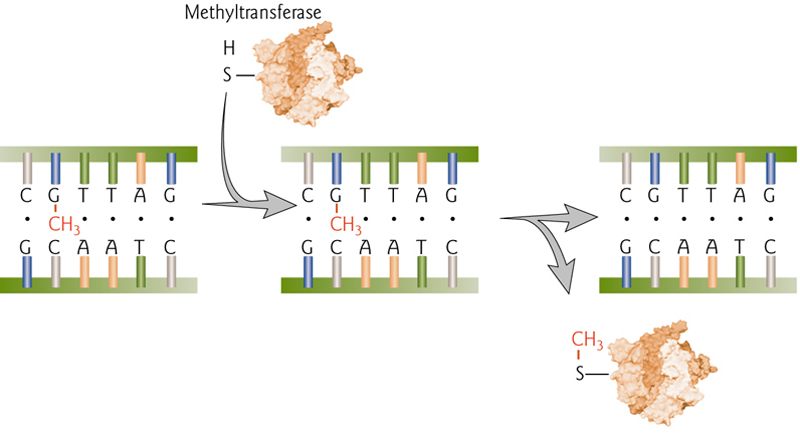
Direct repair of O-6 methylguanasine by permanent transfer to methyltransferase enzyme. This is “costly” to repair because the enzyme is permanently disabled by this modification.
Base Excision Repair
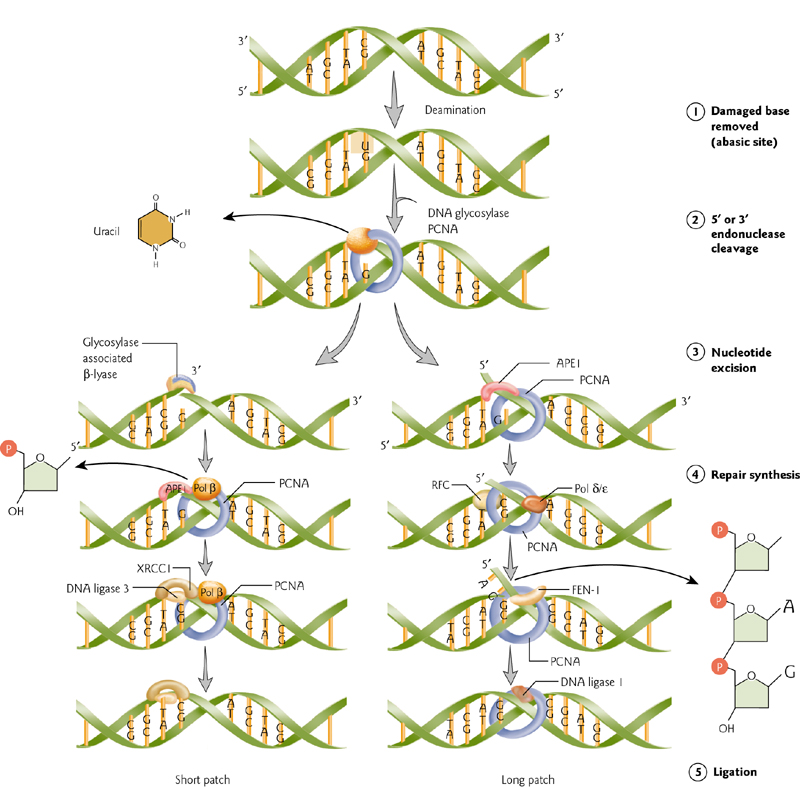
Damaged, single bases can be “swapped out”.
Newly replicated DNA strand is unmethylated, and indicates parent versus daughter strands. Repairs are made to the (new) daughter strand.
Repairs take place by excision of the mismatched base (DNA glycosylase), removal of its baseless deoxyribose phosphate (AP endonuclease), filling-in by DNA polymerase, and sealing of the nick by DNA ligase.
Mismatch Repair
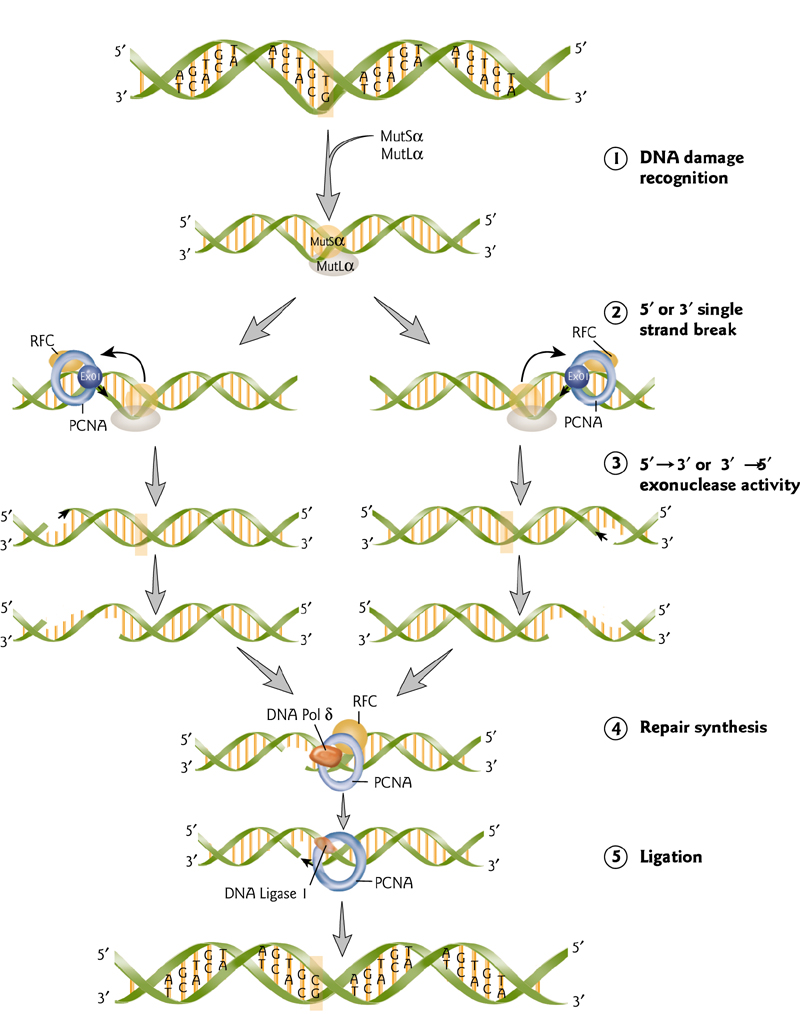
Mismatch repair enzymes are highly conserved through evolution.
Mismatch is recognized and large region of DNA is removed.
This is then filled in and sealed.
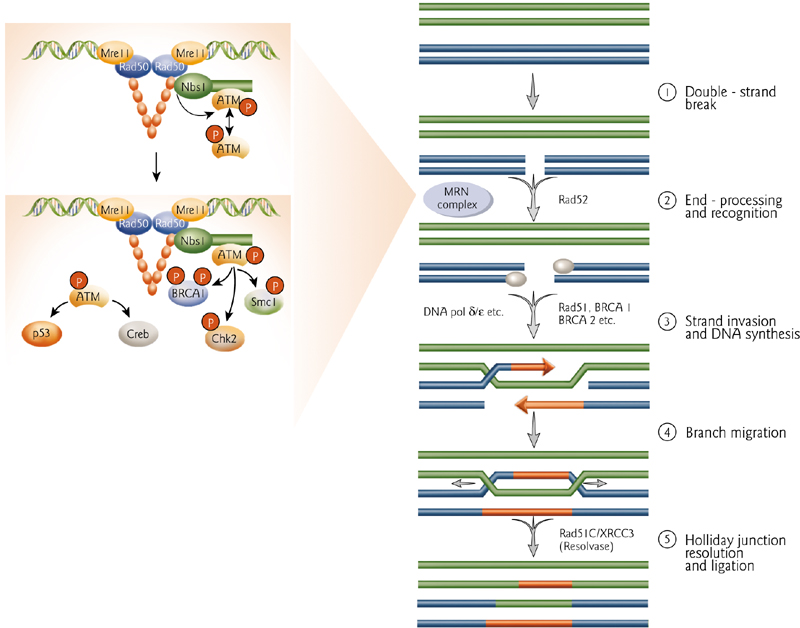
Recombination Repair
1. Honologous Recombination
dsDNA break induced by radiation. MRN complex recruited to break.
Exonuclease activity generates 3’ ssDNA tails that invade homologous undamaged sequences. Missing sequences restored by DNA synthesis. Holliday junctions resolved and repair is completed.
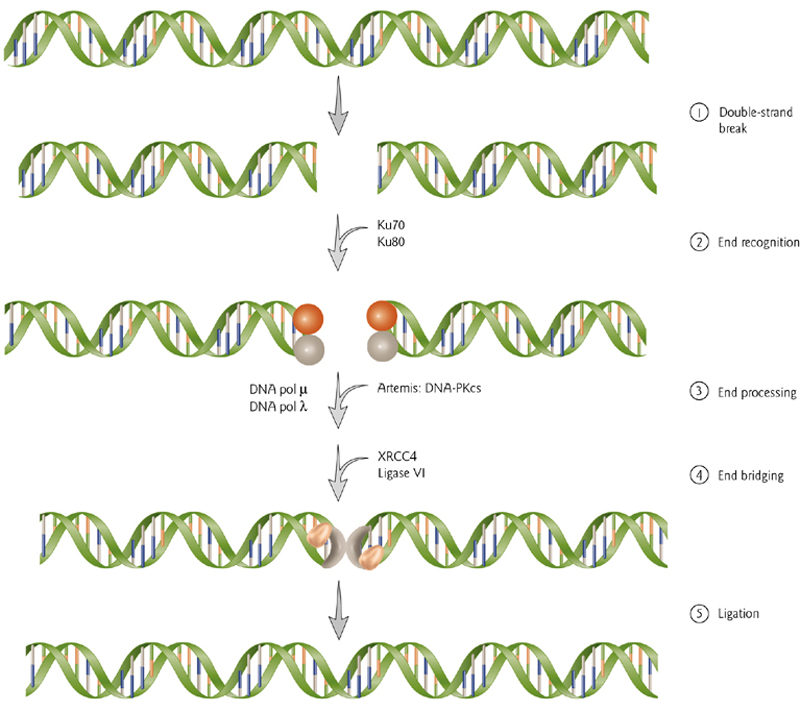
2. Non-Honologous End-Joining
Broken ends are recognized by Ku70/80. An endonuclease (Artemis) trims the ends or damaged nucleotides. DNA polymerase fills it in and ligase complex brings the ends together and completes it.