L7 Restriction enzyme
一、Nomenclature for the restriction Endonuclease
Researcher found some strains of bacteria appeared to be less vulnerable to bacteriophage infections than others and resisted the hijacking of their cell machinery by bacteriophages
In the 1960s, Werner Arber observed a dramatic change in the bacteriophage DNA after it invaded these resistant strains of bacteria: It was degraded and cut into pieces.
How could the host cell distinguish between an invader DNA and its own DNA?
- Arber hypothesized that bacterial cells might express two types of enzymes:
- A restriction enzyme that recognizes and cuts up the foreign bacteriophage DNA.
- A modification enzyme that recognizes and modifies the bacterial DNA to protect it from the DNA-degrading activity of its very own restriction enzyme
This prediction was confirmed in the late 1960s by Stuart Linn and Arber when they isolated a modification enzyme called methylase and a restriction enzyme responsible for bacteriophage resistance in the bacterium Escherichia coli. The methylase enzyme added protective methyl groups to DNA, and the restriction enzyme cut unmethylated (unprotected) DNA at multiple locations along its length.
The restriction modification system (RM system)
- The restriction enzyme cut unmethylated DNA which is exogeneous (from Bacteriophage)
- The methylase enzyme added protective methyl groups to endogenous DNA therefore protect it from degradation by the restriction enzyme.
An overview of Restriction Enzymes
Bacteria have an enzymatic immune system that recognizes and destroys foreign DNA
- Over 10,000 bacteria species have been screened for restriction enzymes.
- Over 3000 have been identified
- More than 600 available commercially
Nomenclature for the Restriction Endonuclease
Named for bacterial genus, species, strain, and type:
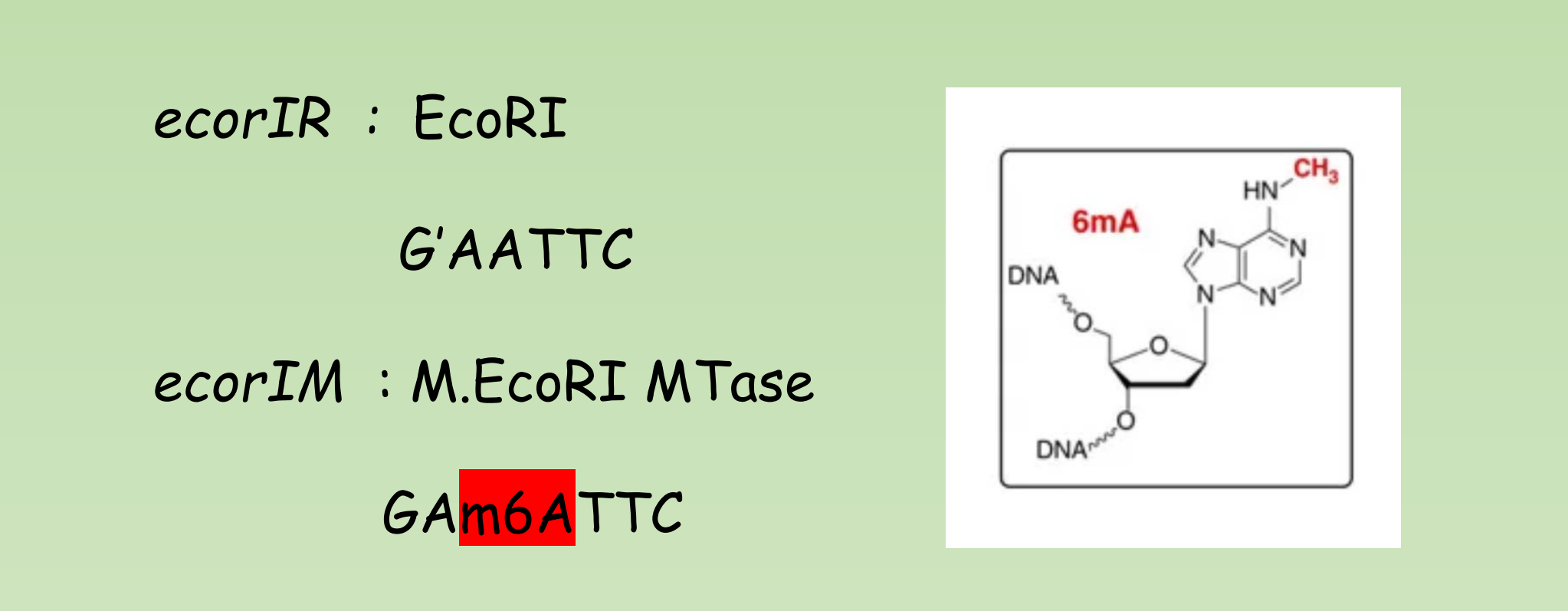
Example: EcoR I
- Genus: Escherichia
- Species: coli
- Strain: R
- Order discovered: 1
1. Different types of Restriction Endonuclease
- Type I and Type III restriction enzymes
Type I- multi-subunit complex, the complex with both endonuclease and methylase activities, cleave at random up to 1000 bp from recognition sequence
Type III- multi-subunit complex, with both endonuclease and methylase activities, cut about 25 bp from recognition sequence
These enzymes are difficult to be used in genetic engineering-multi-subunit, unseparable endonuclease and methylase activities
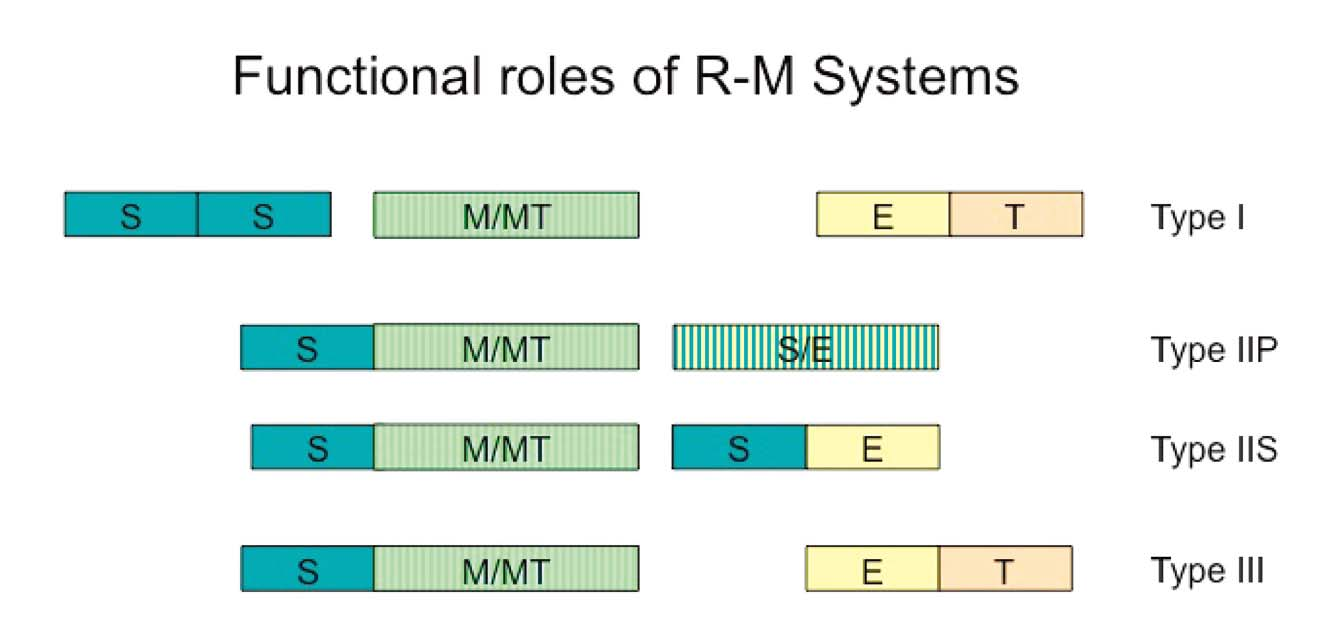
2. Type II restriction enzyme
Type II- the methyltransferase and endonuclease are encoded as two separate proteins and act independently, cleave DNA within or close to the recognition sequence
- Mostly used for gene analysis and cloning
- Recognize 4-8bp sequences
- Need Mg2+ as cofactor
- Cut in close proximity of recognition site
- Homodimer
- ATP hydrolysis is not required
Features of Type II P Restriction Enzyme
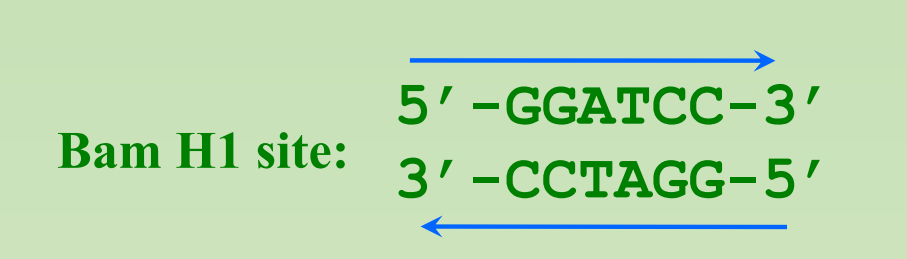
Recognition sites often have symmetry (Palindromic)
- A great bargain: A nut for a jar of tuna
Estimate the frequency of restriction site
How often would a 4bp site of a restriction enzyme appear in the genome?
Recognition sites often 4-8bp long
- You can roughly estimate how frequent a 4bp sequence appears
- Once in 4^4^ = 1/256
Blunt Ends & Sticky Ends
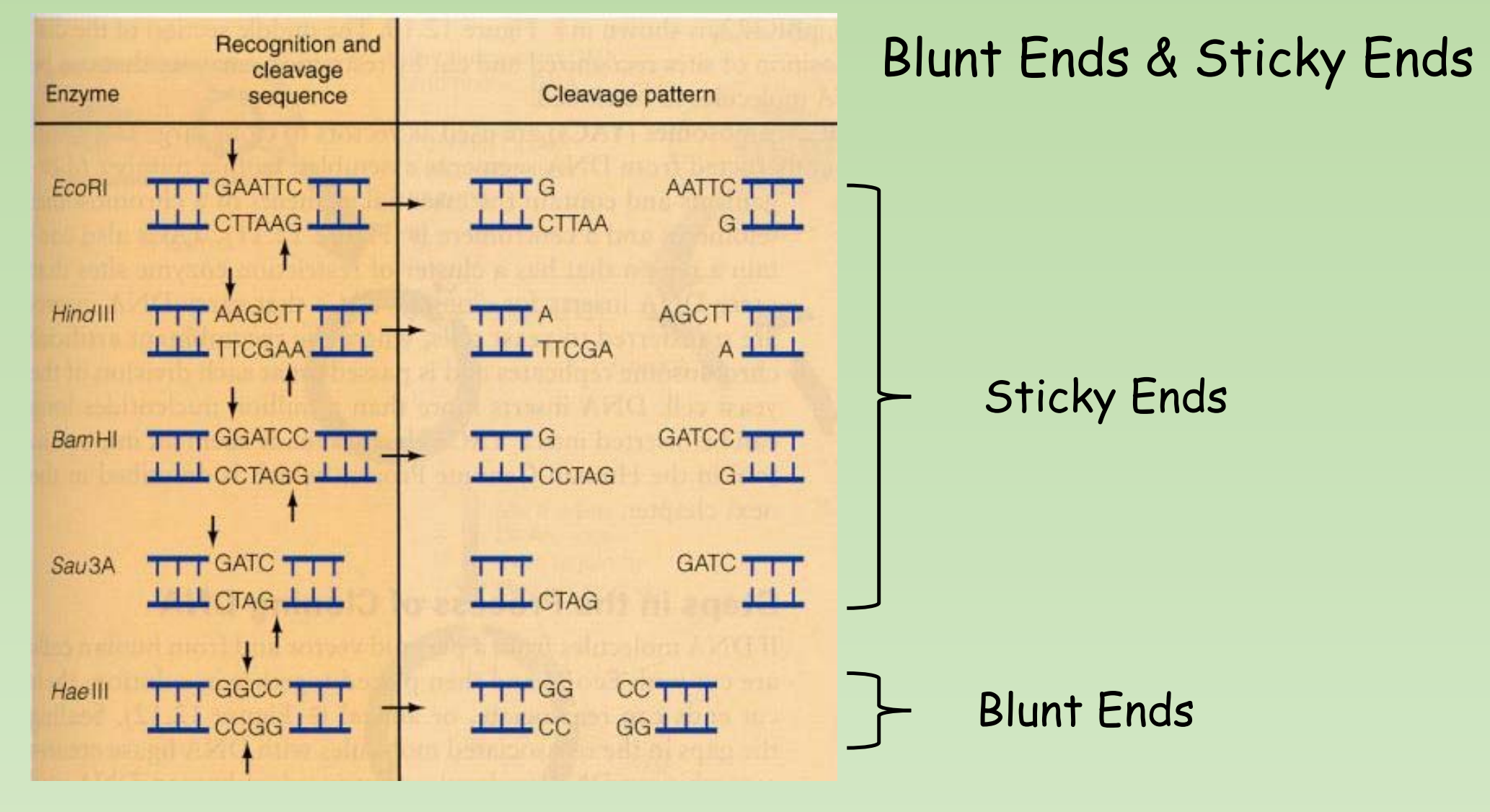
二、Exonuclease & Endonuclease; Sticky End & Blunt End; Star Activity; Isoschizomers & Isocaudamer
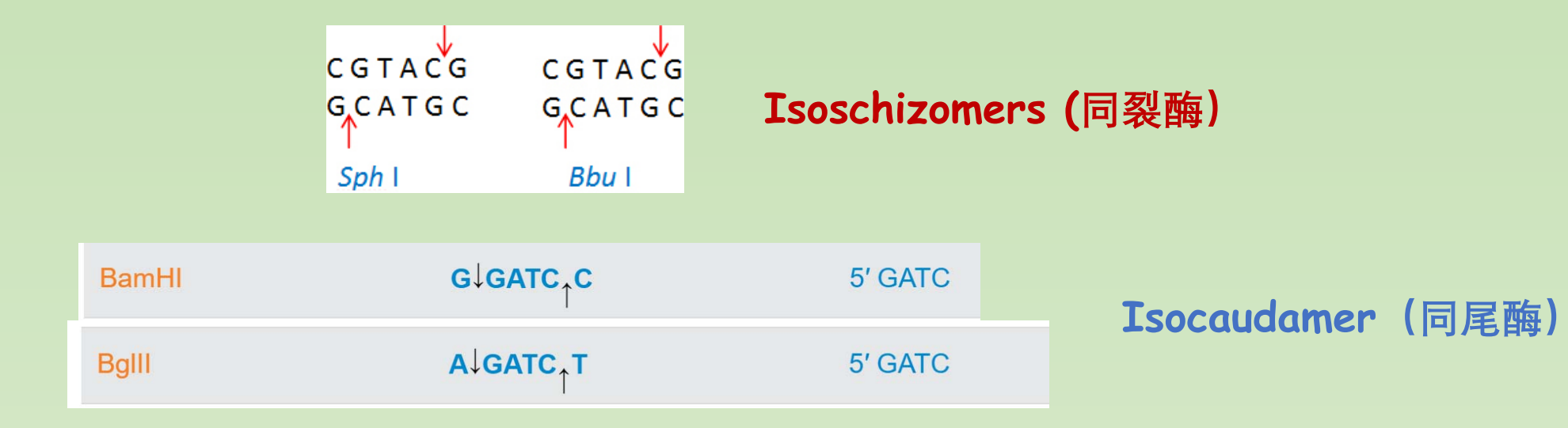
Isoschizomers (同分裂体) are pairs of restriction enzymes specific to the same recognition sequence
- 同一识别序列特有的一对限制性内切酶
An enzyme that recognizes a slightly different sequence, but produces the same ends is an isocaudomer(异分裂体)
- 能识别一个略有不同的序列,但产生相同末端的酶
Star Activity
Star activity is the relaxation or alteration of the specificity of restriction enzyme mediated cleavage of DNA that can occur under reaction conditions that differ significantly from those optimal for the enzyme. – result random pattern
Changed ionic strength, high pH, and high (> 5% v/v) glycerol concentrations can trigger star activity.
Note that commercial restriction enzymes are usually supplied in a buffer containing a substantial amount of glycerol (50% v/v is typical), meaning insufficient dilution of the enzyme solution can cause star activity
三、The application of Restriction Enzyme
Gene Cloning –cut and join two pieces of DNA with the coordination of DNA ligase
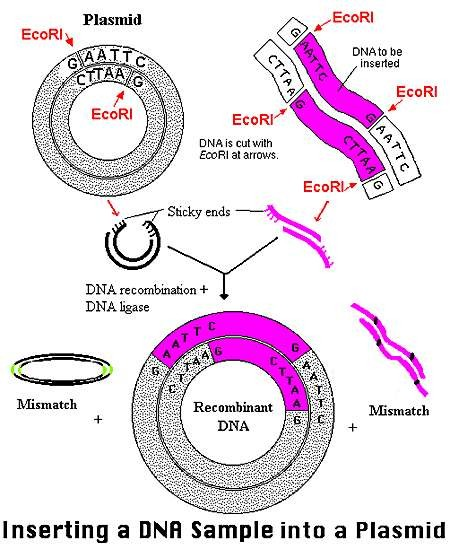
- Designing of primer with restriction site
- Amplifying target gene by PCR and designed primer Double digestion of vector and amplified gene by same REs
- Ligation of gene and vector
- Transformation
- Expression
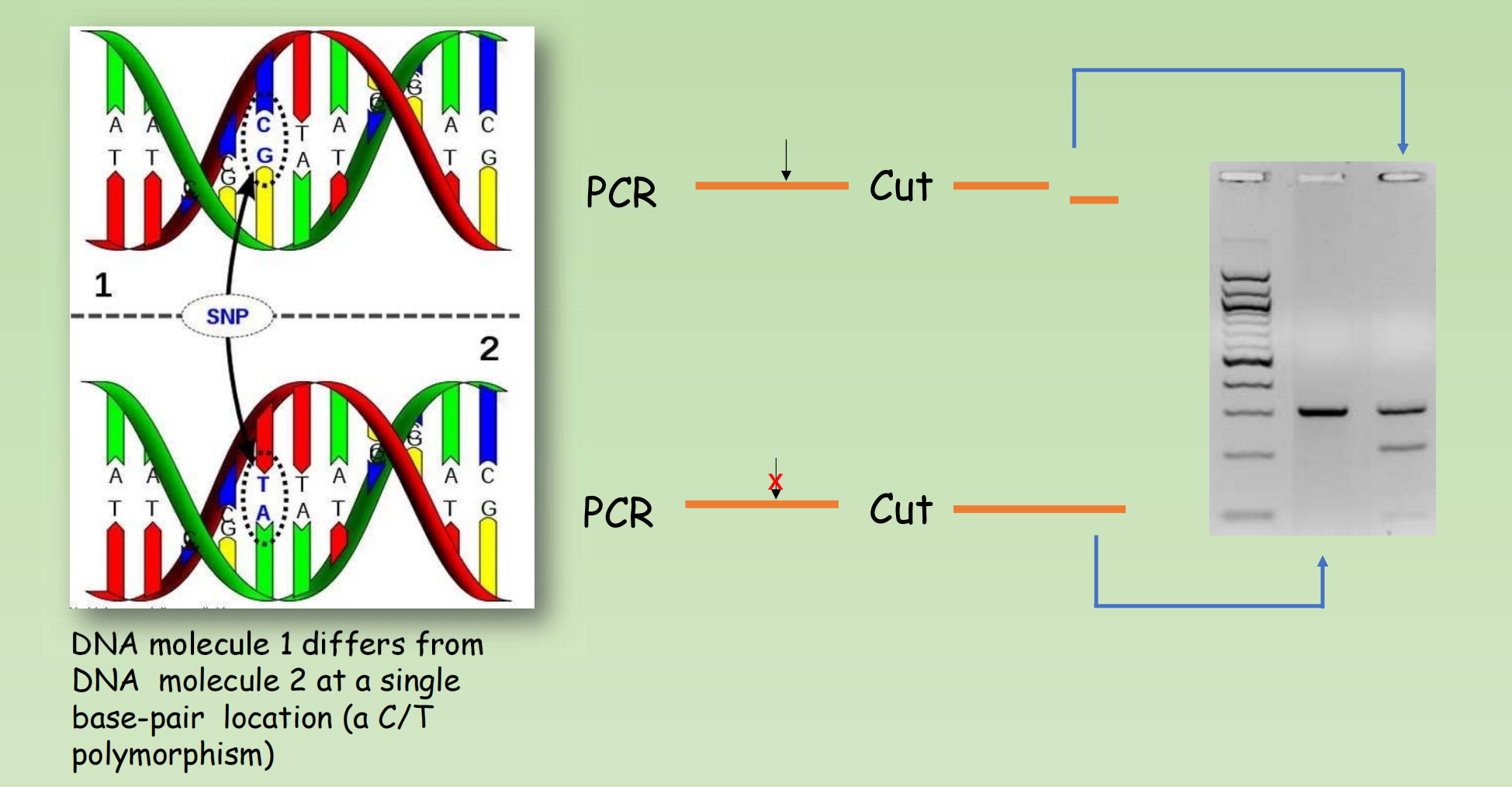
Genotyping a DNA sample – SNP or RFLP
- SNP – Single Nucleotide Polymorphism 单核苷酸多态性
- 单核苷酸多态性主要是指在基因组水平上由单个核苷酸的变异所引起的DNA序列多态性。它是人类可遗传的变异中最常见的一种,占所有已知多态性的90%以上。
- RFLP – Restriction Fragment Length Polymorphism 限制性片段长度多态性
- 限制性片段长度多态性(restriction fragment length polymorphism,RF LP )简称PCR-RFLP 分析。它主要是设计适当的扩增引物,使扩增片段包括一个或数个多态性的限制性内切酶识别序列,在PCR 扩增后用该限制酶切割PCR 产物,根据电泳后酶切(Amp-FLP) 对VNTR.STR 等重复序列,因重复单位数目的不同而呈现高度多态。
Detection the SNP with Restriction Enzyme
RFLP as Molecular Marker
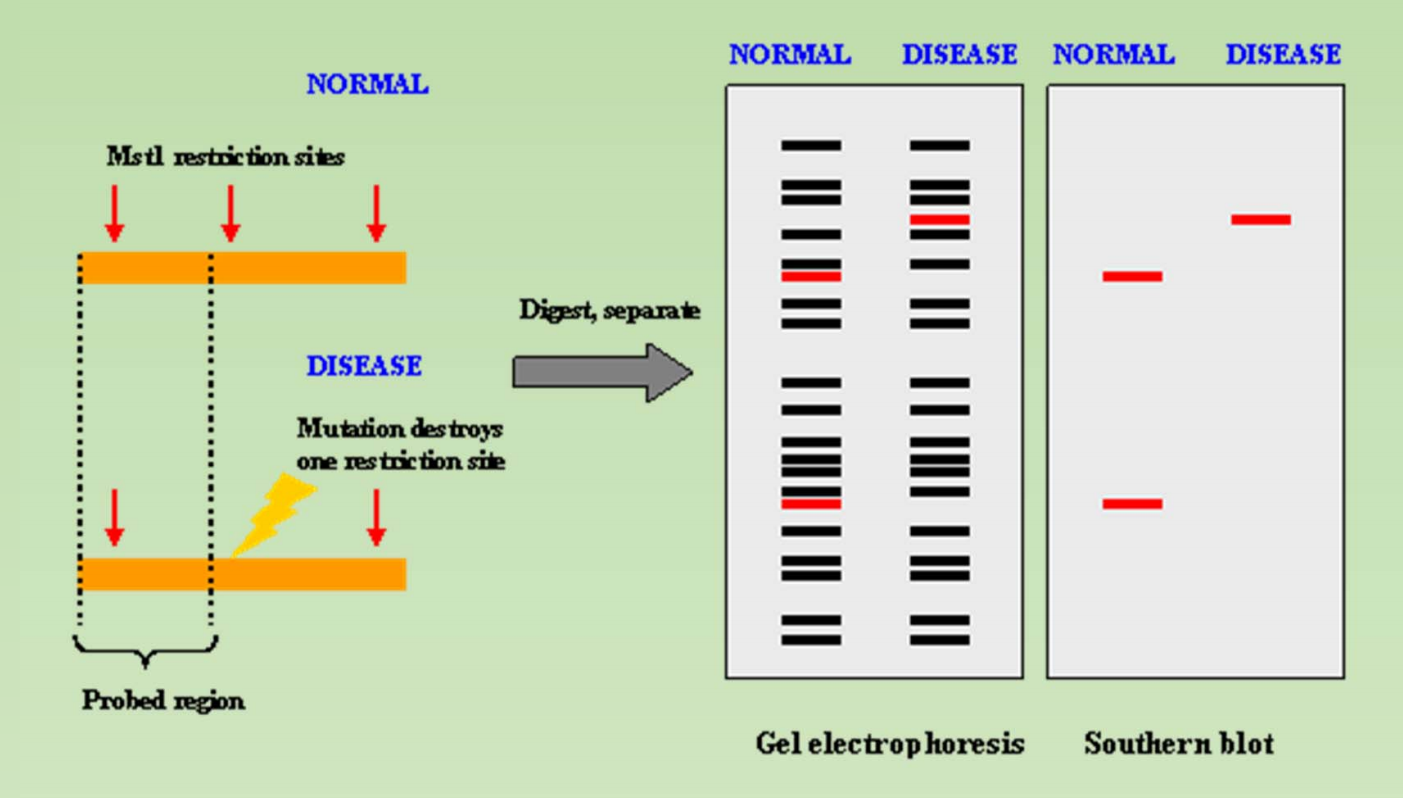
- Analysis of RFLP is used in genome mapping, DNA fingerprinting, genetic disease analysis and identifying mutations (polymorphisms).
