L04 Recombinat DNA
一、Phage
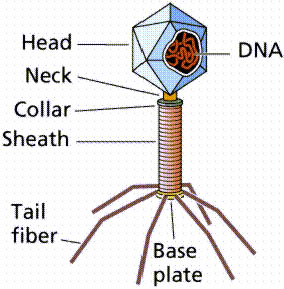
Structure of phages
Phage are relatively simple bacterial parasites that can only reproduce within a bacterial cell. They use ribosomes and other host factors during the stage within the cell, but encode relatively few genes of their own.
Most commonly, phage are linear dsDNA, but can be circular, single stranded, or RNA.
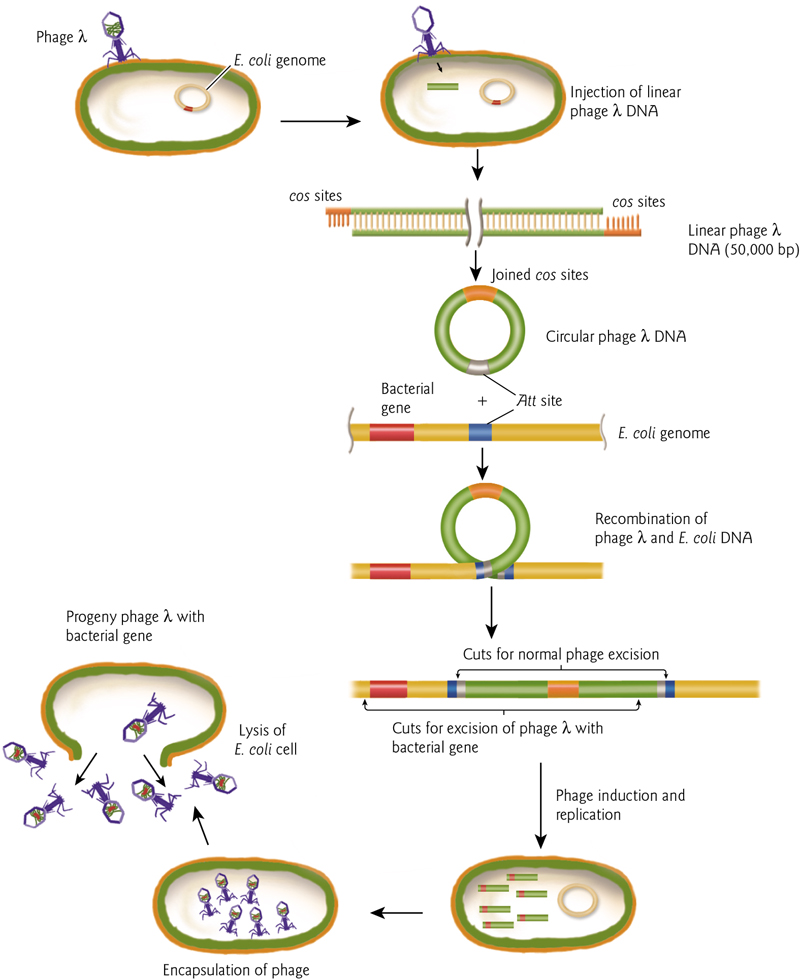
The Life Cycle
- Phage attaches to a specific host bacterium
- injects its DNA
- disrupts the bacterial genome and killing the bacterium
- takes over the bacterial DNA and protein synthesmachinery to make phage parts.
- The process culminates with the assembly of new phage
- the lysis of the bacterial cell wall to release 100’s of new copies of the input phage into the environment.
Restriction and Modification Systems
The bacterial cell uses the restriction enzyme to cut the invading DNA of the phage at the specific recognition site of the enzyme. This prevents the virus from taking over the cellular metabolism for its own replication.
The genes encoding this system are often carried on plasmids, and provide advantages to hosts.
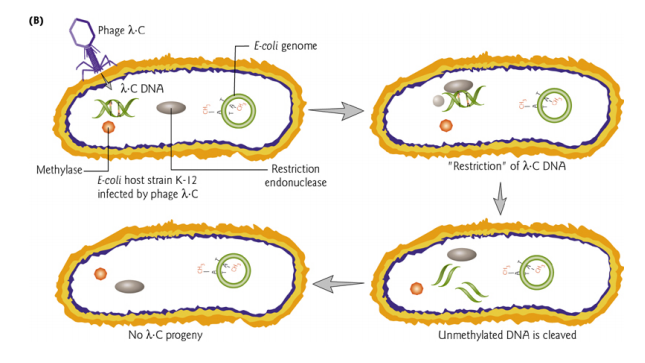
The restriction-modification system is used to protect bacteria from invasion by phage. Bacterial DNA also contains sites cleavable by the restriction enzyme, but is protected by a methylase which recognizes the same target site adds a methyl group to that site.
Methylated sites are not substrates for the restriction enzyme.
The bacterial DNA is methylated immediately following replication so it will not be a suitable substrate for restriction endonuclease cleavage.
1. Nucleases
Nucleases are enzymes that degrade nucleic acids, the opposite function of polymerases.
They hydrolyze, or break, an ester bond in a phosphodiester linkage.
2. Restriction Enzyme
Restriction enzymes are proteins that cut at specific sites
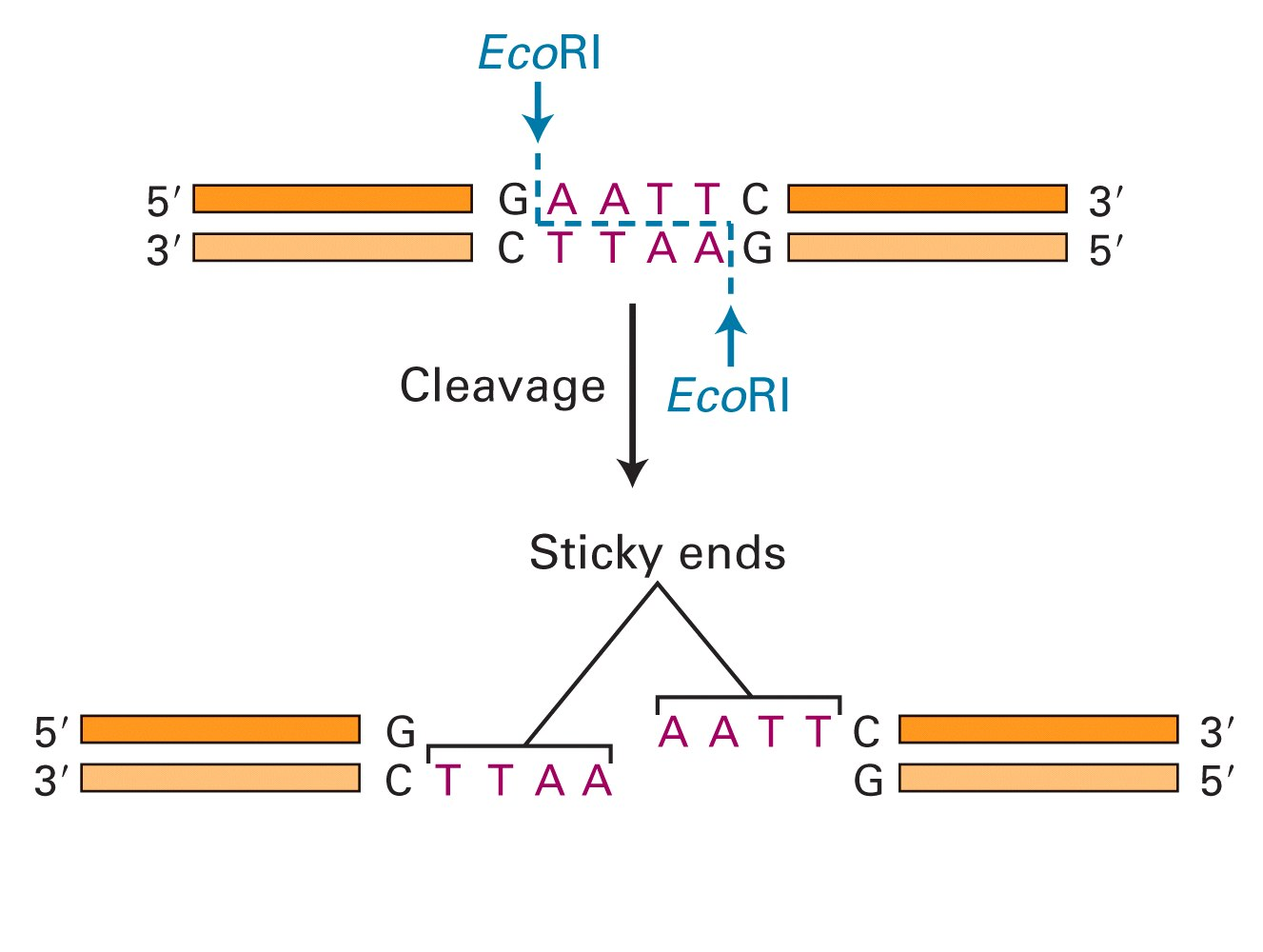
EcoRI was one of the first enzymes isolated. Target sequences areusually short palindromes.(回文的)
These can leave blunt or “sticky” ends, with either 5’ or 3’ overhangs resulting from the symmetrical recognition sites.
Enzymes scan DNA to recognize target. Recognition causes conformational change of the enzyme and DNA, leading to catalysis (cleavage) and DNA release.
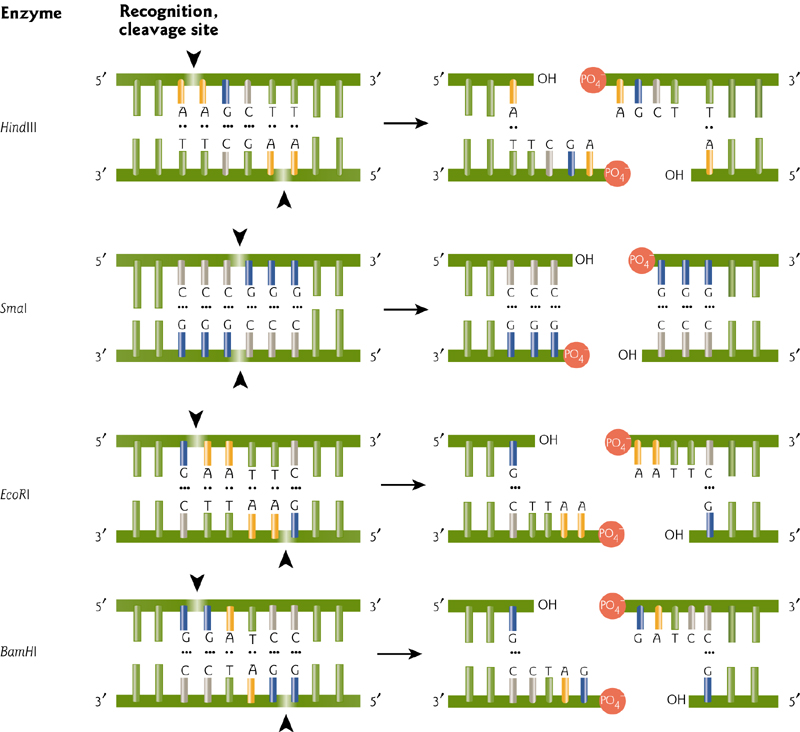
Different enzymes recognize different sequences, usually palindromic (and therefore 4, 6, or even 8 bp recognition sites…why an even number?).
These can leave blunt or “sticky” ends, with either 5’ or 3’ overhangs resulting from the symmetrical recognition sites.
The 1978 Nobel prize was awarded for the discovery and use of restriction enzymes.
Steps Involved In Cleavage By Type II Restriction Enzyme
Enzymes scan DNA to recognize target. Recognition causes conformational change of the enzyme and DNA, leading to catalysis (cleavage) and DNA release.
3. Ligase
Ligases are used to seal cuts in the DNA strands.
A “vector(载体)” is a molecule that can replicate in a host cell-like a small circle of DNA in a bacterial cell.
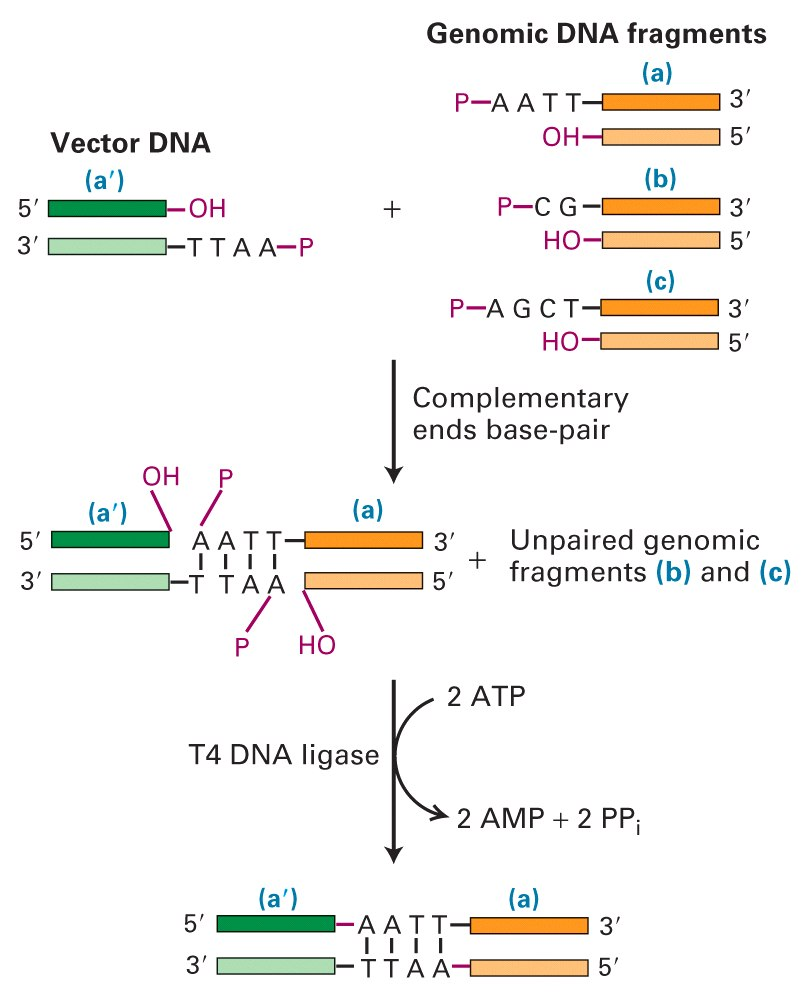
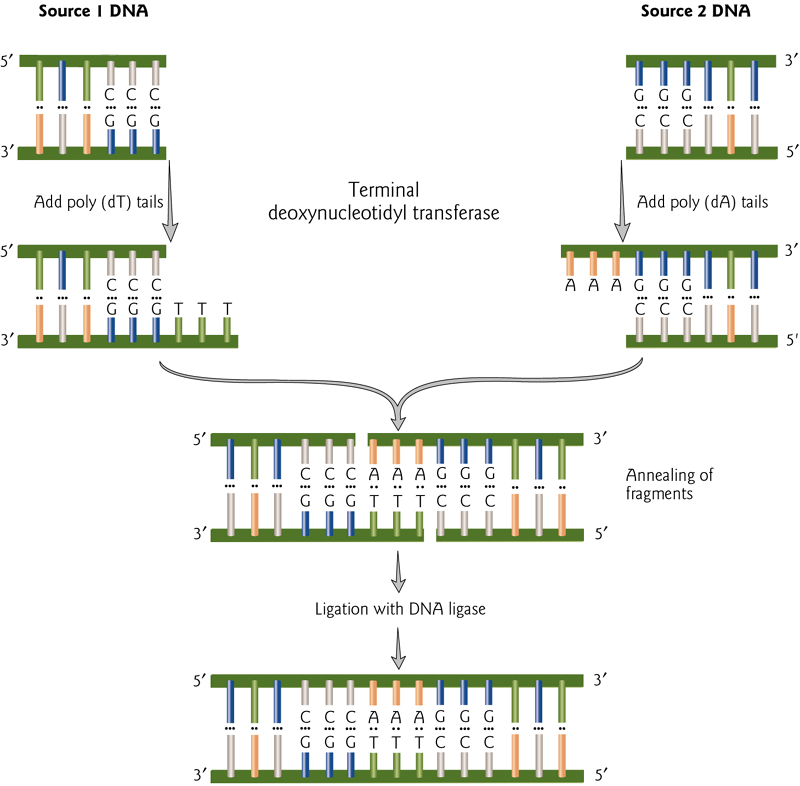
Blunt End Ligations(平末端)
Ligation of blunt ends has efficiency, but can be increased by adding complementary tails of poly(dA) and poly(dT).
These can then be covalently linked by DNA ligase.
二、Techniques about Recombination DNA
Vectors
| Vector | Features | Isolation of DNA | DNA limit |
|---|---|---|---|
| Plasmid | High copy number | Physical | 10 kb |
| Phage | Infects bacteria | Via phage packaging | 20 kb |
| Cosmid | High copy number | Via phage packaging | 48 kb |
| BAG | Based on F plasmid | Physical | 300 kb |
| YAC | Origin + centromere + telomere | Physical | 3000 kb |
- Plasmid 质粒 - an extrachromosomal circular DNA molecule that autonomously replicates inside the bacterial cell; cloning limit: 100 base pairs to 10 kb
- Phage 噬菌体- derivatives of bacteriophage lambda; linear DNA molecules, whose region can be replaced with foreign DNA without disrupting its life cycle; cloning limit: 8 to 20 kb
- Cosmids 黏质体、粘粒- an extrachromosomal circular DNA molecule that combines features of plasmids and phage; cloning limit: 35 to 50 kb
- Bacterial Artificial Chromosomes (BAC,细菌人工染色体) - based on bacterial mini-F plasmids. cloning limit: 75 to 200 kb
- Yeast Artificial Chromosomes (YAC,酵母人工染色体) - an artificial chromosome that contains telomeres, origin of replication, a yeast centromere, and a selectable marker for identification in yeast cells; cloning limit: 100 to 1000 kb
1. Plasmids
A plasmid is an independent, circular, self-replicating DNA molecule that carries only a few genes, and these have been found in many organisms (bacteria to plants and animals).
The number of plasmids in a cell generally remains constant from generation to generation.
Plasmids are autonomous molecules and exist in cells as extrachromosomal genomes, although some plasmids can be inserted into a bacterial chromosome, where they become a permanent part of the bacterial genome.
These molecules often confer selective advantage, but may be genetically dispensible.
Plasmids provide great functionality in molecular biology.
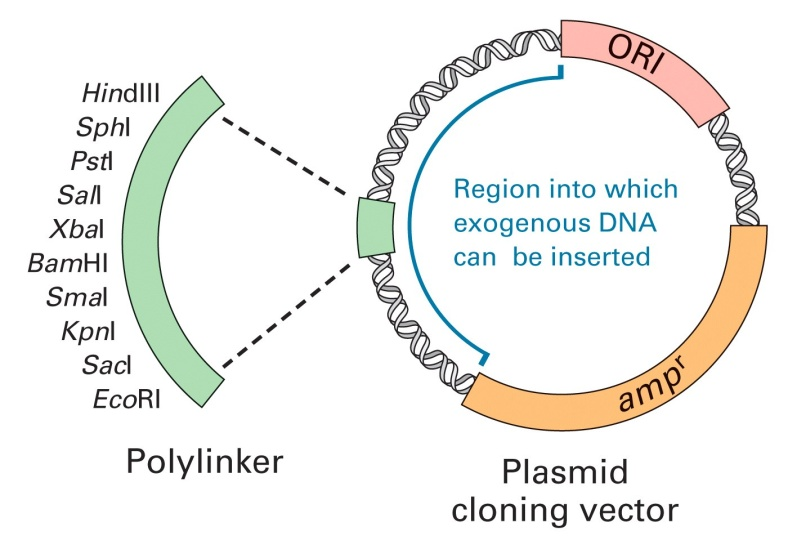
Plasmid structure: usually negatively supercoiled circular dsDNA.
Naturally occurring plasmids were named for phenotypes they conferred (F for fertility, R for antibiotic resistance, etc.)
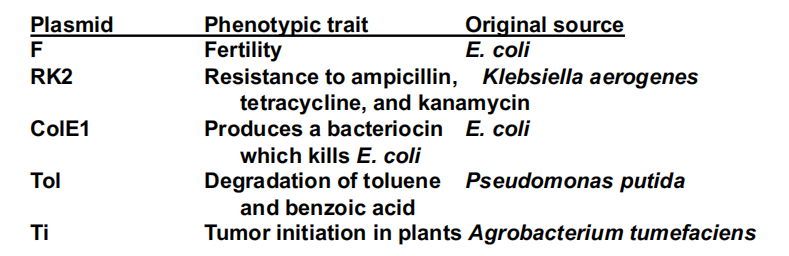
Artificially constructed plasmids are usually given the designation “p” for plasmid, “XY” the initials of the person or team that constructed the plasmid, and “nnn,” its serial number. Thus pBR322 was the serial number 322 plasmid constructed by Bolivar and Rodriquez.
Use Plasmids to do molecular cloning
Vector + DNA fragment → Recombinant DNA → Replication of recombinant DNA within host cells → Isolation, sequencing, and manipulation of purified DNA fragment
Marker Genes
In nuclear biology and molecular biology, a marker gene is a gene used to determine if a nucleic acid sequence has been successfully inserted into an organism’s DNA. In particular, there are two sub-types of these marker genes: a selectable marker and a marker for screening.
Plasmids that express the lacZ 5’ end can be used in E. coli expressing the lacZ 3’ end.
The protein fragments form a functional enzyme that cleaves the colorless X-gal (5- bromo-4-chloro-3- indolyl- beta-D-galactoside) into a blue colored product.
2. Phage
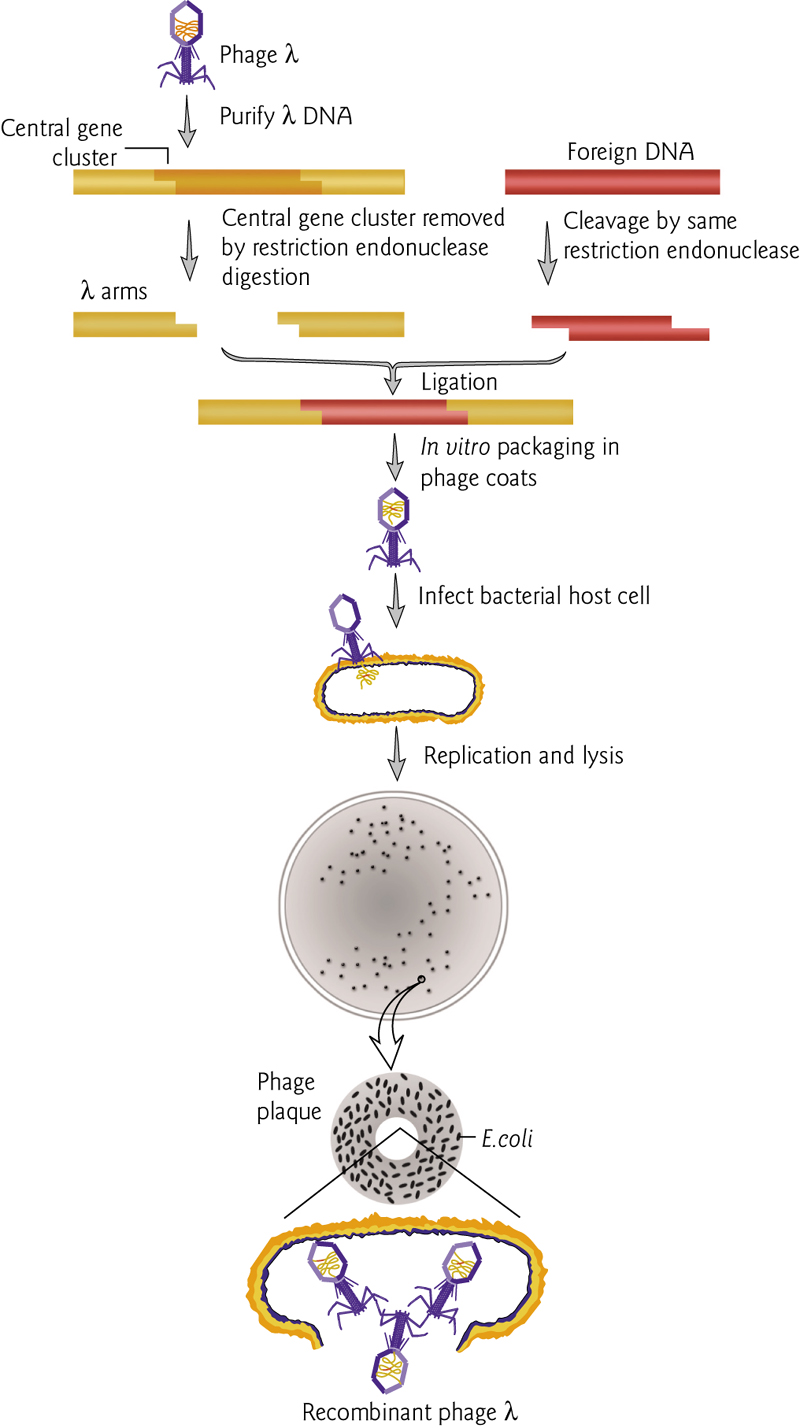
Bacteriophage Lambda as a cloning vector:
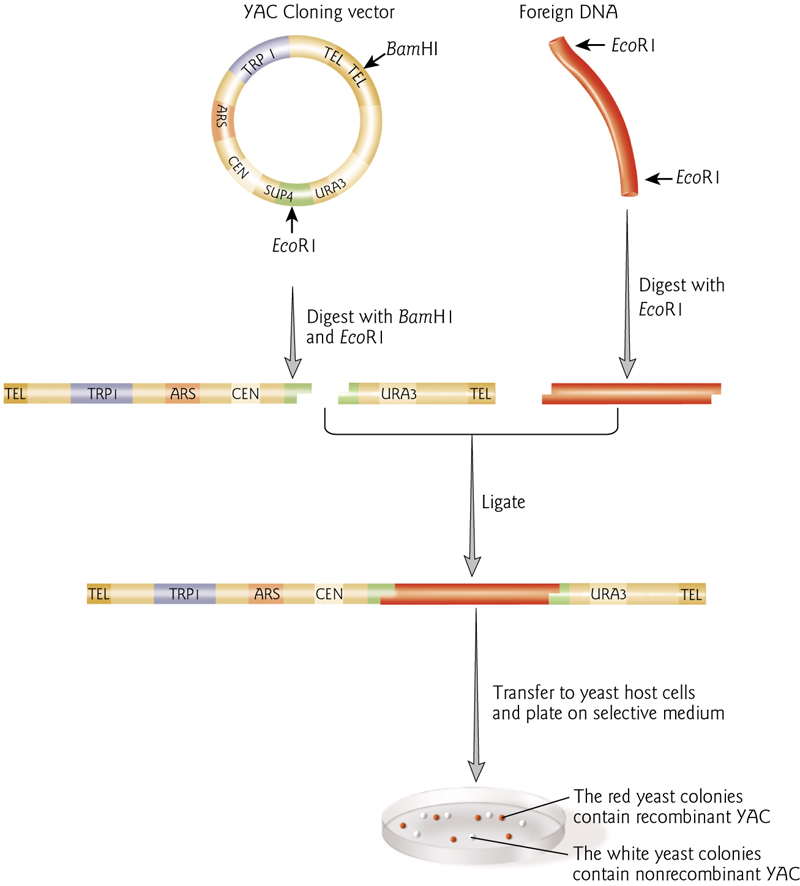
3. YAC
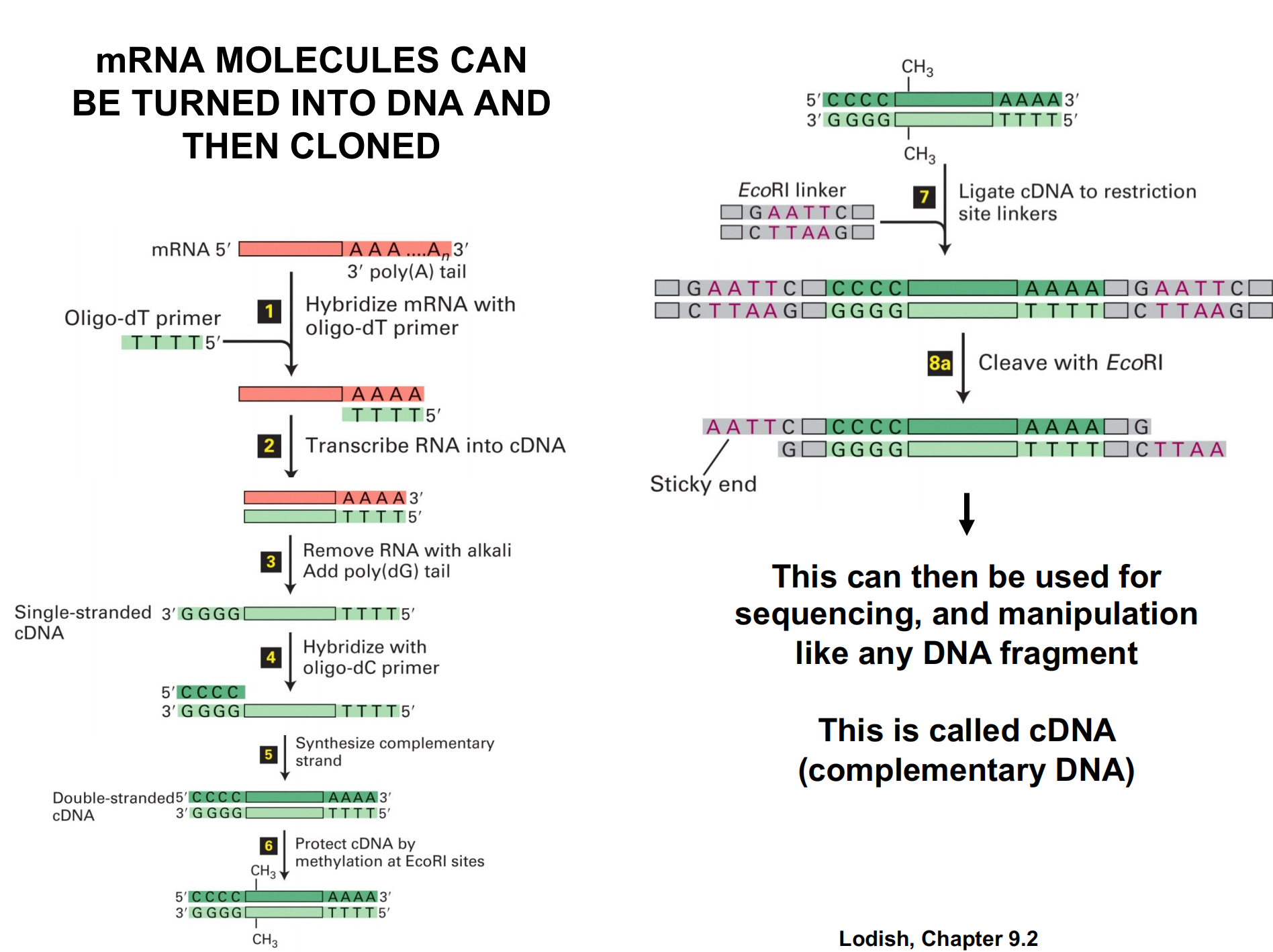
mRNA can be turned to DNA and the cloned
“Gateway” Cloning
Use site specific recombinases
Probes and Labeling of Molecules
Searching for a particular DNA sequence in a library of molecules requires a specific “probe”.
“labels” to track the probe can be added to the end (“end-labelled”) or incorporated internally during replication
DNA probes can be made from synthetic oligonucleotides, cloned DNA fragments, or RNA probes (riboprobes).
DNA must be denatured prior to use as a probe, but RNA is already single-stranded.
Heterologous probes are similar to the target but not an exact match. Homologous probes are exactly complementary.
Methods Labeling Nucleic Acids
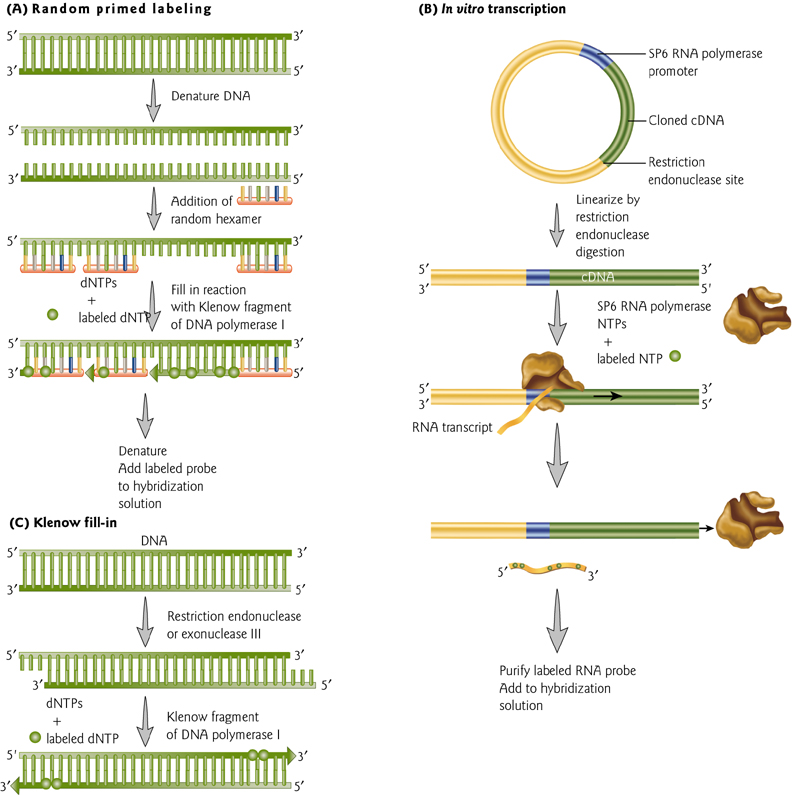
Two methods of uniform labeling and one method of end labeling – all require nucleic acid synthesis.
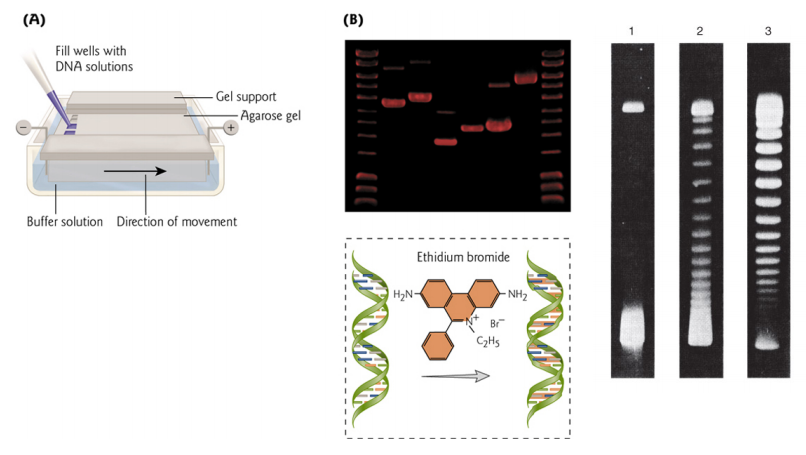
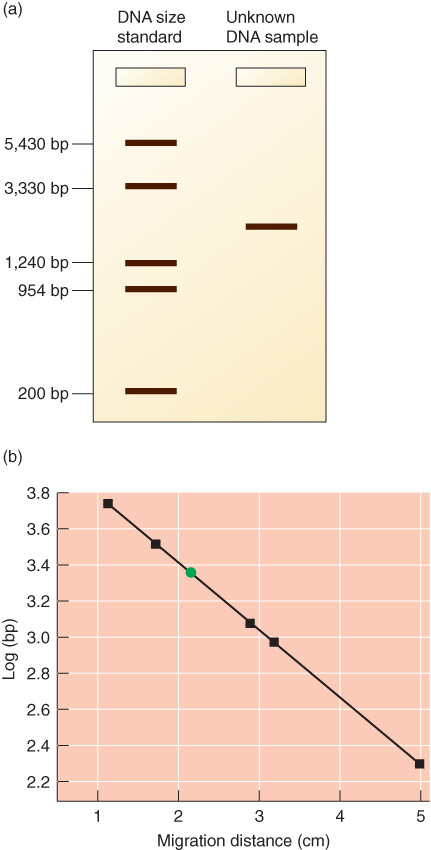
Agarose Gel Electrophoresis
DNA size can be determined by Gel Electrophoresis
1. Southern Blots
DNA is separated, transferred to a nylon membrane (maintaining the relative position of fragments in the agarose gel).
The size of a specific piece of DNA is checked using a labelled probe.
Steps:
- Digest ONA with restriction endonucleases
- Perform agarose gel electrophoresis on the DNA fragments from different digests
- DNA fragments fractionated by size (visible under UV light if gel is soaked in ethidium bromide)
- Transfer (blot) gel to nitrocellulose or nylon membrane using Southern blot technique
- DNA fragments are bound to the membrane in positions identical to those on the gel
- Hybridize membrane with radioactively labeled probe
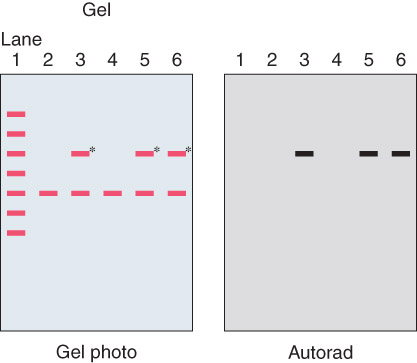
- Expose membrane to X-ray film. Resulting autoradiograph shows hybridized DNA fragments
Autoradiograph 自体放射造影照片
An autoradiograph of a gel to check for specific sequences
Western Blot
西方墨点法(英语:Western blot)或称「蛋白质转渍法」、「免疫墨点法」(immunoblot)或「西式吸印杂交」,是分子生物学、生物化学和免疫遗传学中常用的一种实验方法,也是HIV检测的方法之一。
利用特定抗体能够专一结合其抗原蛋白质的原理来对样品进行着色,通过分析着色的位置和着色深度获得特定蛋白质在所分析的细胞或组织中的表达情况的信息,来分析检测特定蛋白质的生物学检测技术。
Western Blot与Southern或Northern杂交方法类似,但Western Blot采用的是聚丙烯酰胺凝胶电泳,被检测物是蛋白质,“探针”是抗体,“显色”用标记的二抗。经过PAGE分离的蛋白质样品,转移到固相载体(例如硝酸纤维素薄膜)上,固相载体以非共价键形式吸附蛋白质,且能保持电泳分离的多肽类型及其生物学活性不变。以固相载体上的蛋白质或多肽作为抗原,与对应的抗体起免疫反应,再与酶或同位素标记的第二抗体起反应,经过底物显色或放射自显影以检测电泳分离的特异性目的基因表达的蛋白成分。该技术也广泛应用于检测蛋白水平的表达。