L4 Qualification and Quantification of DNA&RNA
一、Handling and Quantification of DNA/RNA

- Pure DNA/RNA only form tiny pellet after precipitation. - semi transparent and easy to be dissolved
Mainly two types of quantification methods
- Absorbance based methods
- the principle is simple, the key is the ratio.
- Fluorescence based methods
The Absorbance Based Methods
The Beer‐Lambert Law — draws a direct correlation between absorbance and concentration. While nucleic acids absorb at many wavelengths, they have a peak absorbance of UV light at 260nm. Thus, the amount of light absorbed in this region can be used to determine the concentration of RNA or DNA in solution
$$
A = \varepsilon c l
$$
- A=absorbance,
- ԑ=extinction coefficient
- c=concentration
- l=path length
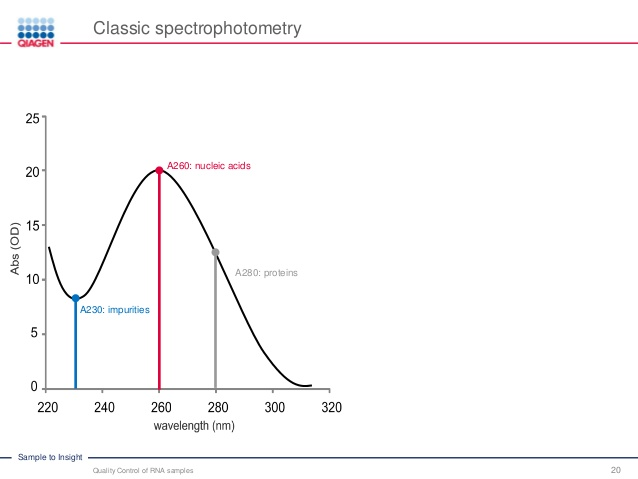
- The nucleic acids (DNA, RNA and oligo) solutions with different concentration has different ability absorbing light.
- 1 OD
260Unit $\rarr$ 50ug/ml for double stranded DNA - 1 OD
260Unit $\rarr$ 40ug/ml for single stranded RNA - 1 OD
260Unit $\rarr$ 33ug/ml for single stranded DNA (ssDNA)
- 1 OD
1. The Ideal Ratios
What would be the ideal ratios for pure DNA and or RNA in solution?
A260/280 ratio
- A low A260/A280 ratio often indicate protein contamination, Aromatic amino acid side chains are responsible for 280nm absorbance.
- For DNA: A260/280 is nearly 1.8
- For RNA: A260/280 is nearly 2.
- Reason: RNA will typically have a higher 260/280 ratio due to the higher ratio of Uracil compared to that of Thymine
A260/230 ratio, ideally more than 1.8
- The A260/230 ratio indicates the presence of organic contaminants which could interfere with downstream applications
- Low A260/A230 often indicate presence of following impurities
- Plant polyphenol – common contaminants when doing RNA/DNA extraction from plant tissues.
- Phenol
- Chaotropic salts from your lysis buffer
2. Real World Examples
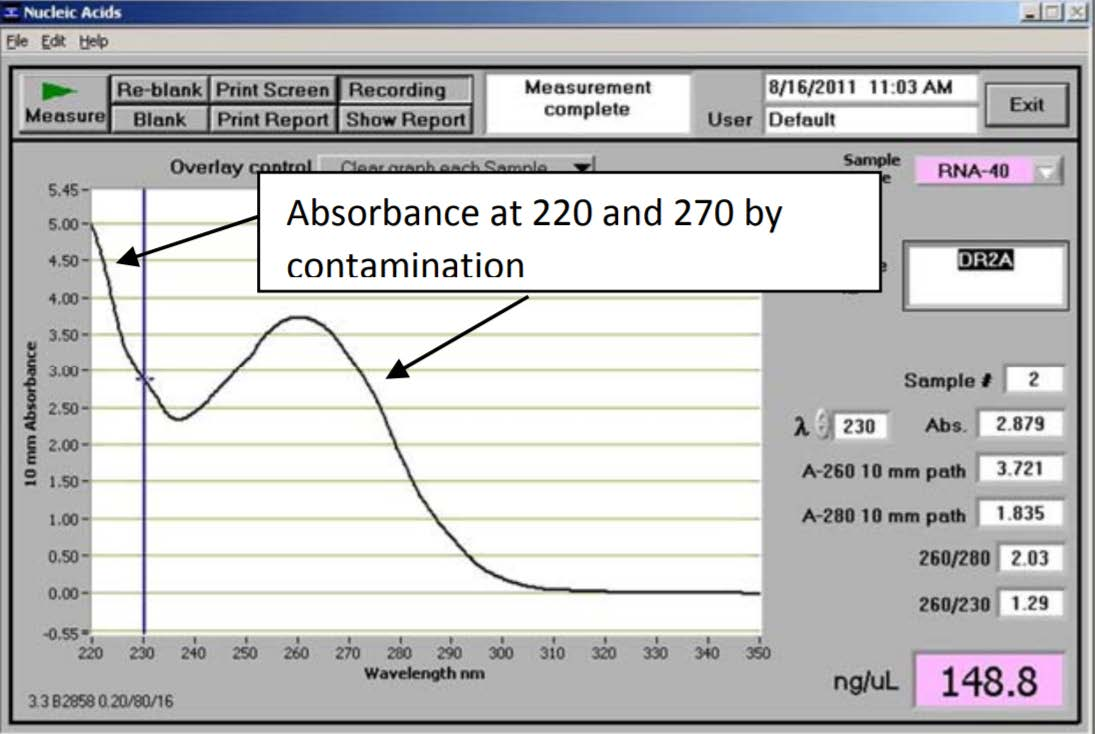
- the A260/230 ratio of 1.29 is significantly lower than 2—indicating some sort of organic contaminants present in the sample

- This RNA has low A260/280 and A260/230 ratios. Additionally, the maximum peak is at 270 nm. Most likely due to presence of high concentration of phenol
- Note that the concentration of RNA returned by the spectrophotometer is not at all reliable due to the contamination

3. How to Keep The Ratio Constant
- Constant ionic strength – same buffer as blank and dilution
- Constant pH – pH 8.0 1xTE buffer is recommanded
Fluorescence Based Methods
1. The shortage of absorbance based method
- Can’t distinguish between DNA/RNA/free nucleotide
- Might not be accurate when concentration of DNA/RNA is low, due to the presence of other impurities.
- No size information
- Might not be sensitive – below 10ng/ul
2. Stains
Fluorescent DNA/RNA Stains– Bind nucleic acids and emit fluorescence when excited.
- DNA(RNA) Stains are fluorescent dyes that bind nucleic acids and have a wide range of applications, including in flow cytometry, cell-cycle studies, chromosome and nuclei counterstaining, as an indicator of apoptosis, and to quantify DNA
The technology involved:
- Gel electrophoresis
- DNA or RNA specific dye enables accurate quantification
- Capillary electrophoresis + DNA/RNA specific dye
Gel Electrophoresis

- agarose gel electrophoresis (SAGE)
- Polyacrylamide-based gel electrophoresis (PAGE)
Gel electrophoresis is a typical fluorescence based methods
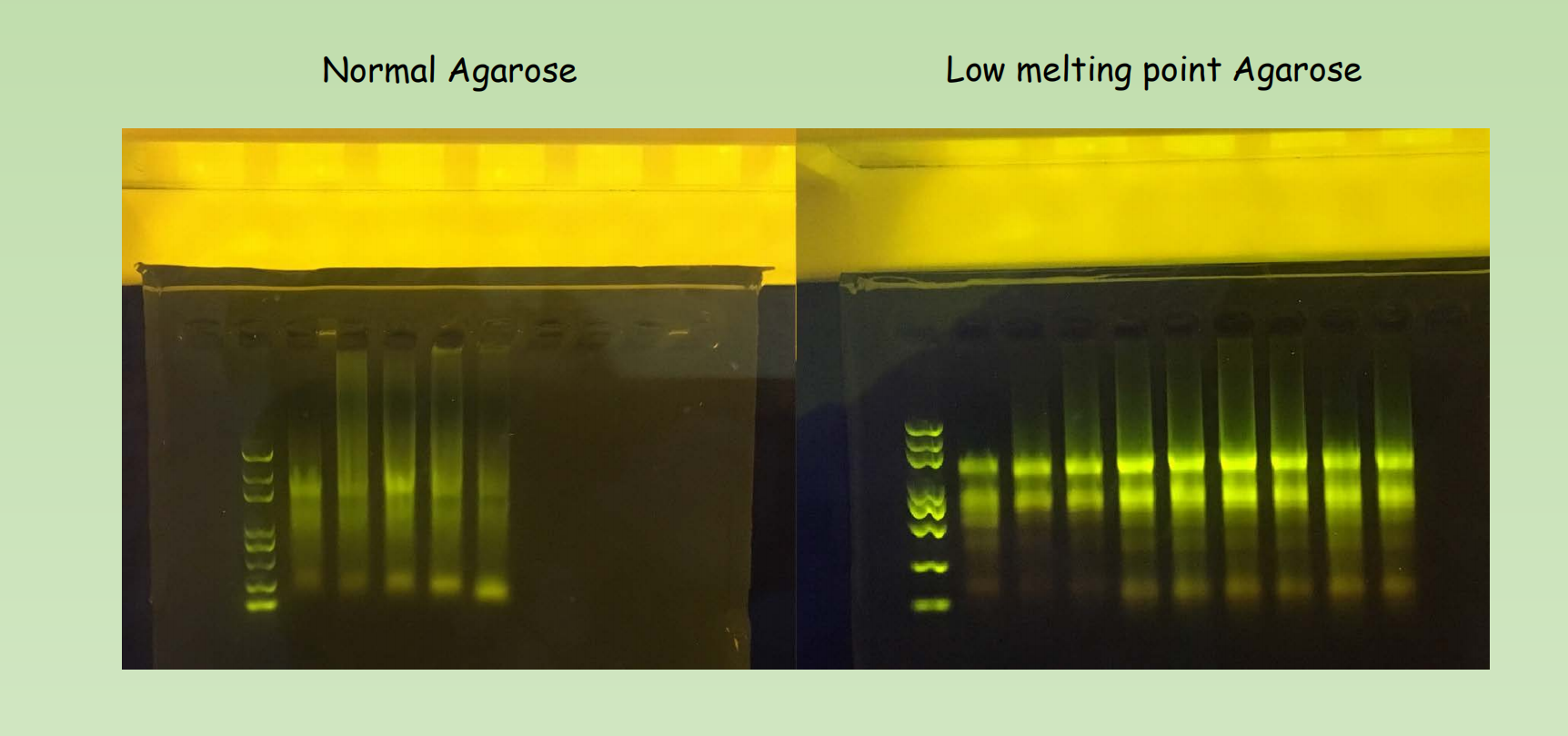
- Matrix of choice – Agarose vs Low-melting point agarose, Polyacrylamide
- Buffer of choice – TAE vs TBE buffer
The migration of DNA fragments through an agarose gel

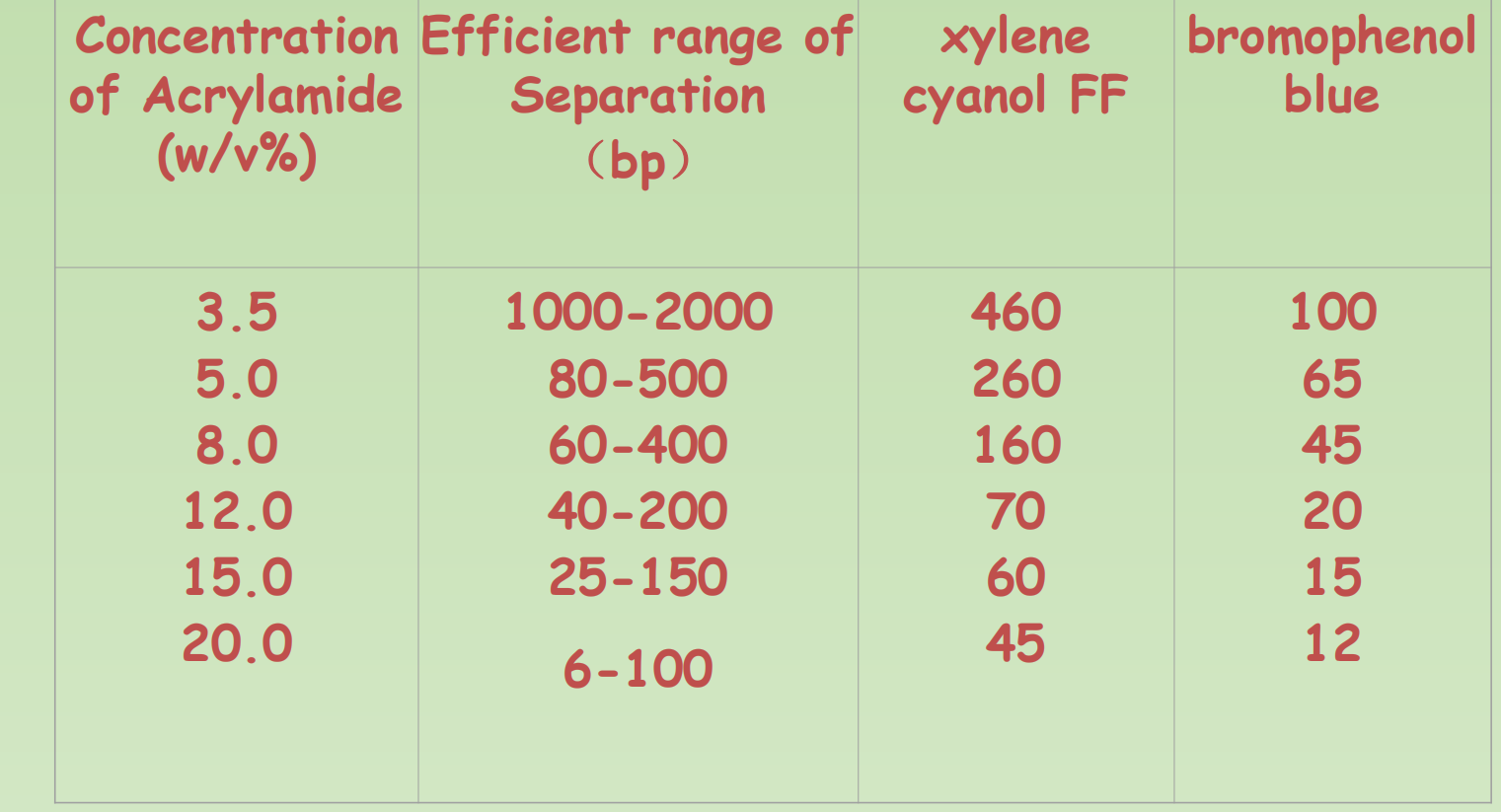
Polyacrylamide Gel Electrophoresis
- 在制备的过程中,加入了cross-linker
- 因此制造出了more ordered struacture,同时也更难配置

Migration of dsDNA in gels with different concentration of acrylamide
Gel electrophoresis is a typical fluorescence based methods.
- Matrix of choice – Agarose vs Low-melting point agarose, Polyacrylamide
- Agarose gel electrophoresis (SAGE) – Work well for a wide range of DNA length. Maxima resolution is around 10bp, High Capacity.
- Low-melting-point agarose, you may choose it for various reason. It is a lot more transparent and have more even pore size compared with normal agarose gel, so it is a better matrix for the detection of DNA/RNA in low concentration
- Polyacrylamide-based gel electrophoresis (PAGE) – High resolution, 1bp resolution is possible. Low Capacity. Good for separate small RNAs and used in DNA sequencing.
- Denatured and Native
Buffer of Choice
Buffer of choice – TAE vs TBE buffer
- TAE is advantageous for high resolution of long nucleic acid fragments (qs) on agarose gels.
- TBE has a greater buffering capacity and will give sharper resolution than TAE, especially when separating small fragments of DNA.
Stains
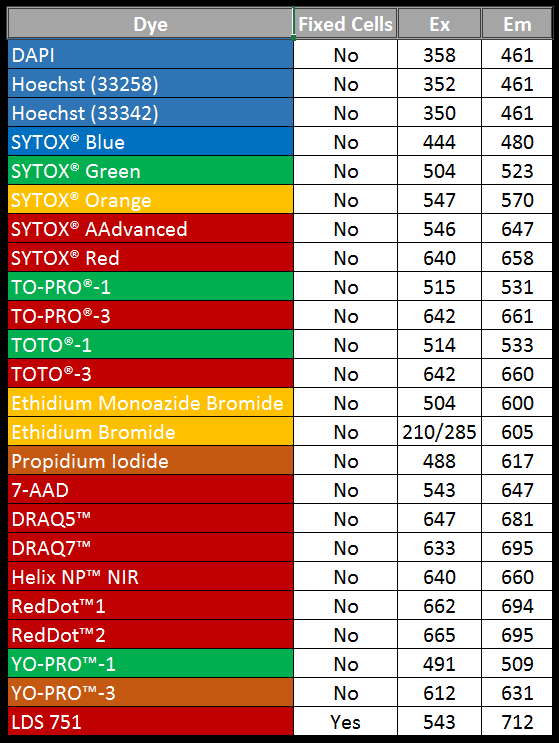
Fluorescent DNA/RNA Stains– Bind nucleic acids and emit fluorescence when excited
We often use two types of DNA stains in gel electrophoresis and other applications.
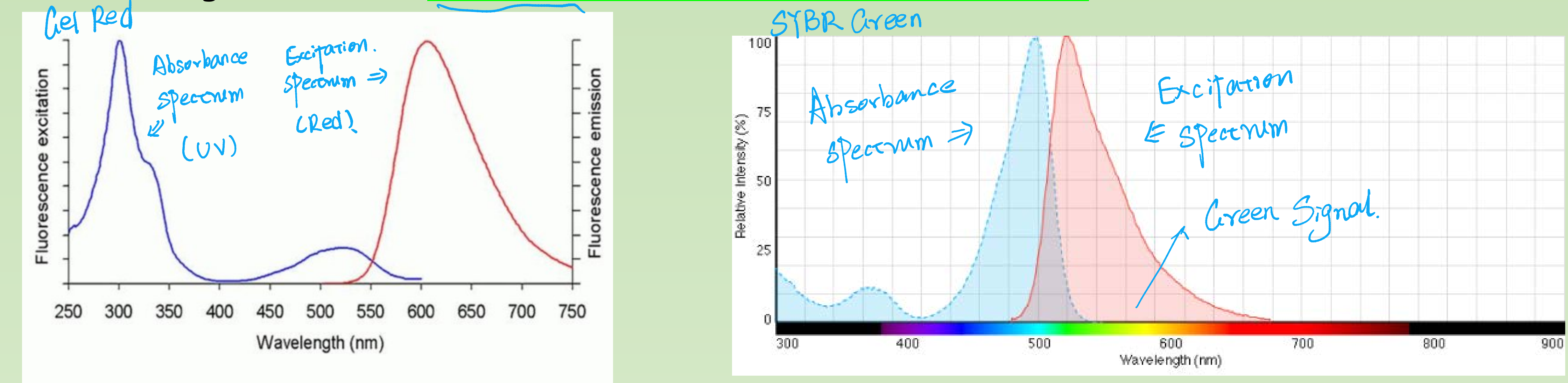
- UV light excited - Ethidium bromide, Gel Red,
- Blue light excited - SYBR Green, Gene Green, Gel Green etc
DNA or RNA specific fluorescent Dye enables accurate quantification of DNA or RNA

The Qubit 1X dsDNA HS (High-Sensitivity) Assay Kit is designed specifically for use with the Qubit Fluorometer. The assay is highly selective for double-stranded DNA (dsDNA) over RNA and is designed to be accurate for initial sample concentrations from 10 pg/µL to 100 ng/µL.


Advantages:
- accurate – highly selective
- sensitive – can deal with sample of very low concentration
Disadvantages:
- Good for quantification but not for qualification (length, integrity).
- Concentration rely on external standard
Capillary electrophoresis + DNA/RNA specific dye
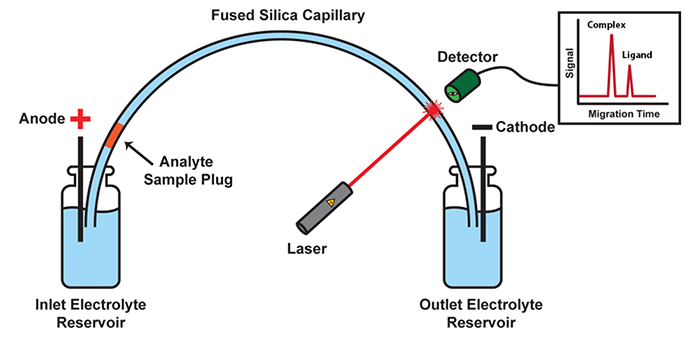
Advantages: You get almost all the information you need !
Disadvantages: Operation with the instrument can be complicated, Expensive (both instrument and reagent)
二、Detection of Nucleic acids through blotting techniques (Nucleic acids hybridization)
Nucleic Acid Hybridization: Detection of nucleic acids with known sequence from complex samples !
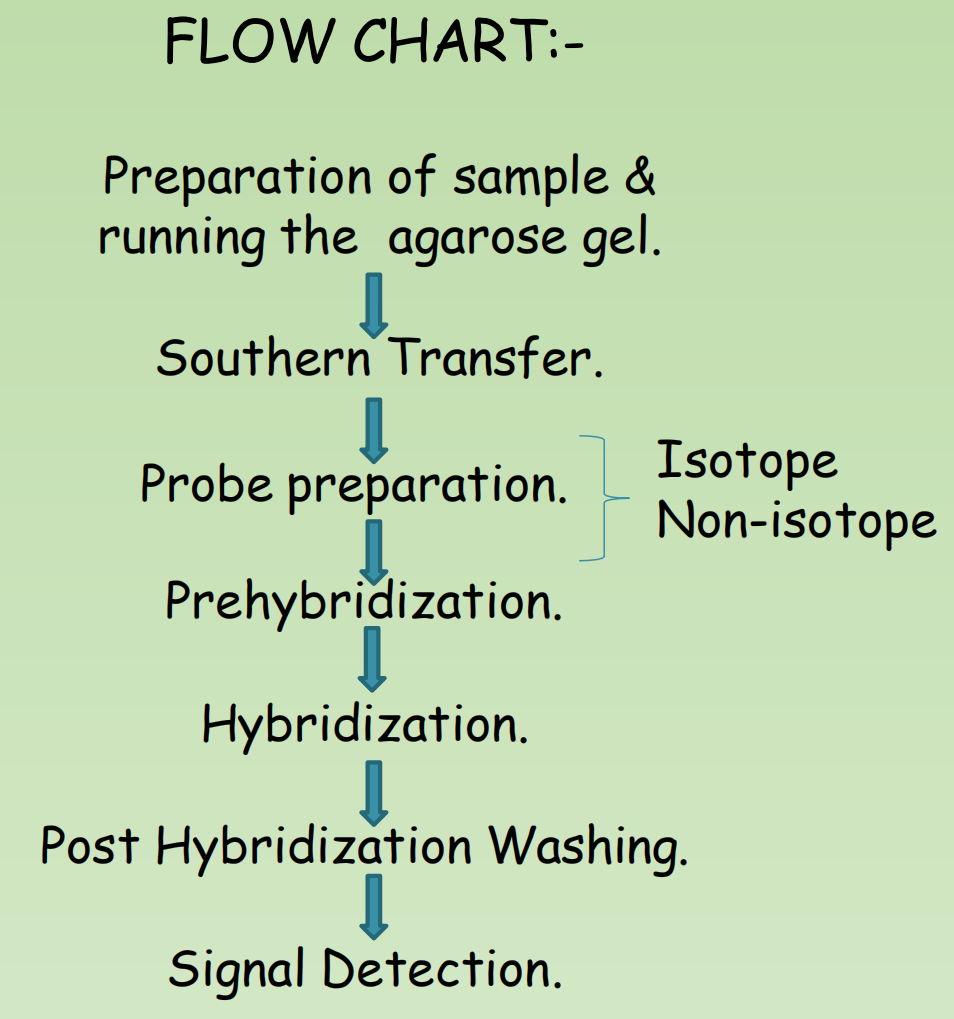
Introduction
A basic technique in molecular biology. in which single stranded Nucleic acids are allowed to interact so that complexes called HYBRIDS are formed by molecules with similar complementary sequences
The essence of nucleic acids hybridization is the denaturation and renaturation (annealing) of DNA (or RNA).
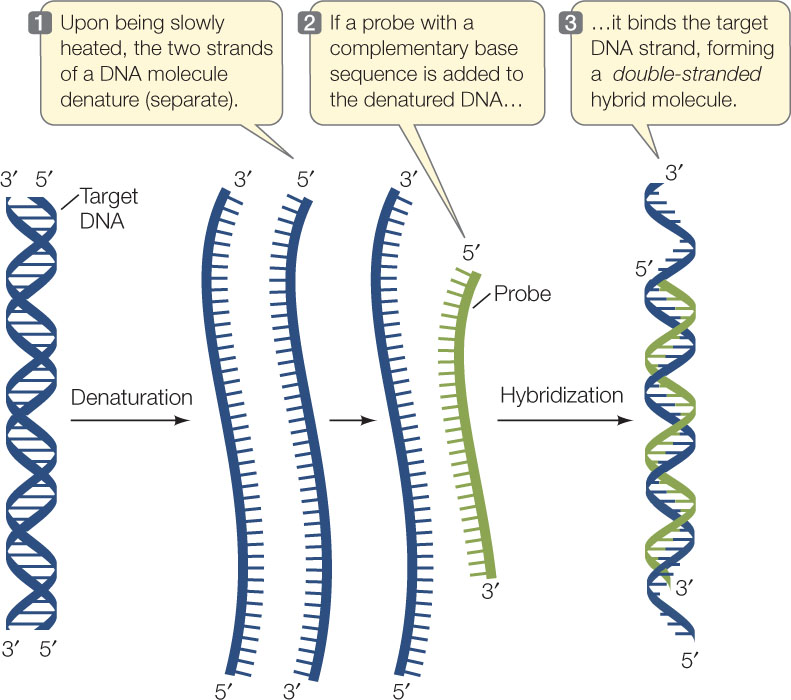
The principle of hybridization is the addition of a probe to a complex mixture of target DNA. The mixture is incubated under conditions that promote the formation of hydrogen bonds between complementary strands.
Different Blotting
Nucleic acid hybridization – A brief history of Southern/Northern/Western blot
- In 1975 when Ed Southern invented his method of using a radiolabeled DNA probe to detect a specific DNA sequence within a DNA sample (e.g. a fractionated genome) and named it after himself – the Southern blot
- In 1977, J.C. Alwine, a biologist with a sense of humor, developed Northern Blot, a technique analogous to the Southern blot, this time for the identification of a specific RNA within a complex RNA sample using a radio-labelled DNA probe.
- W. Neal Burnette, a post-doc working in the Nowinski group at the Hutchinson Cancer Center in Seattle, developed a technique call western
- Seattle sitting in the west cost of US. If it is in New York, it is another story perhaps.
- Burnette was searching for a way to combine the powers of radio immunoassay and SDS-PAGE electrophoresis so that he could pinpoint specific antigens in a complex protein mixture
1. Southern Blotting
- The first type of blotting to be discovered
- Plays significant role in Recombinant DNA Technology as well as Molecular Biology.
- Used to detect target DNA in a sample
Usage of Southern blot: To detect a specific DNA sequence within a complex DNA sample.
- Detection of copy number of a certain gene
- Genotyping – different restriction patterns between two alleles of a same gene
- Detection of replication intermediate – 2D gel + southern blot.
Detection of an RFLP (Restriction fragment length polymorphism) by Southern Blotting:
In molecular biology, restriction fragment length polymorphism (RFLP) is a technique that exploits variations in homologous DNA sequences, known as polymorphisms, in order to distinguish individuals, populations, or species or to pinpoint the locations of genes within a sequence.
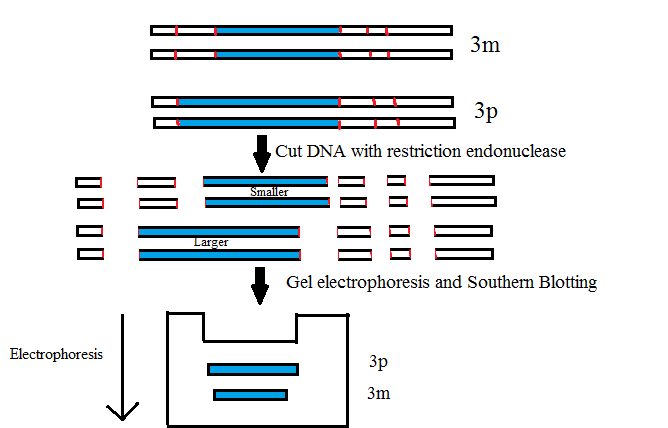
Detection of DNA defect & DNA segment (Gene Defect) (Detection of the sickle cell globin gene)
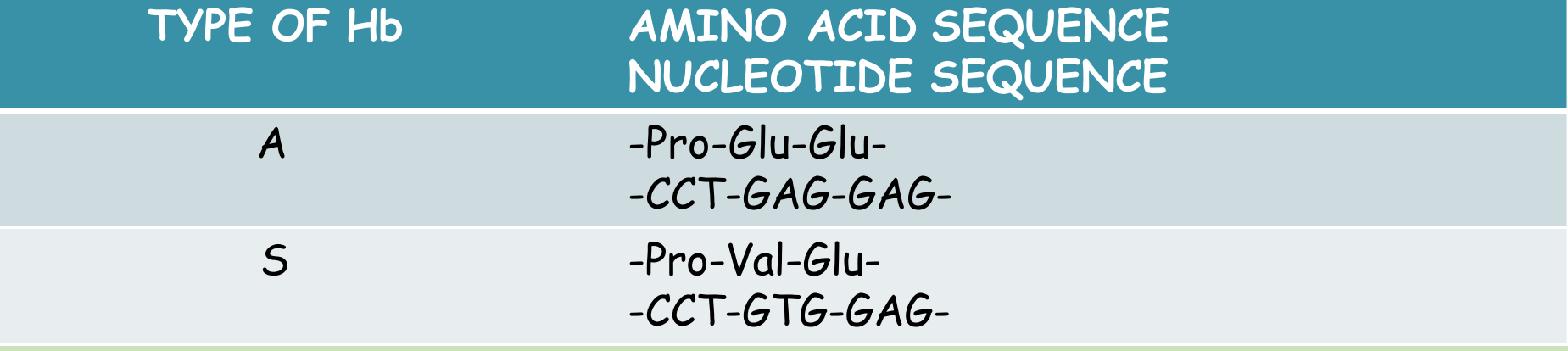
- Using restriction enzymes we can prove that they are different types of genes.
- Normal DNA- 2 segments
- Sickle cell- 1

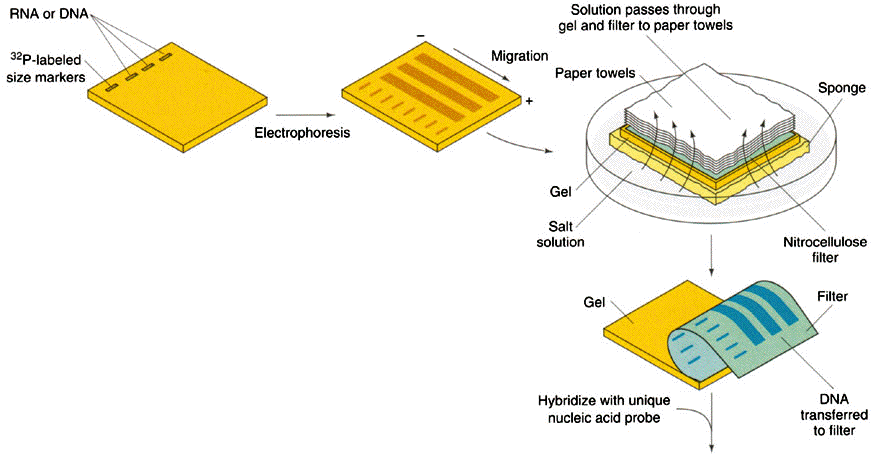

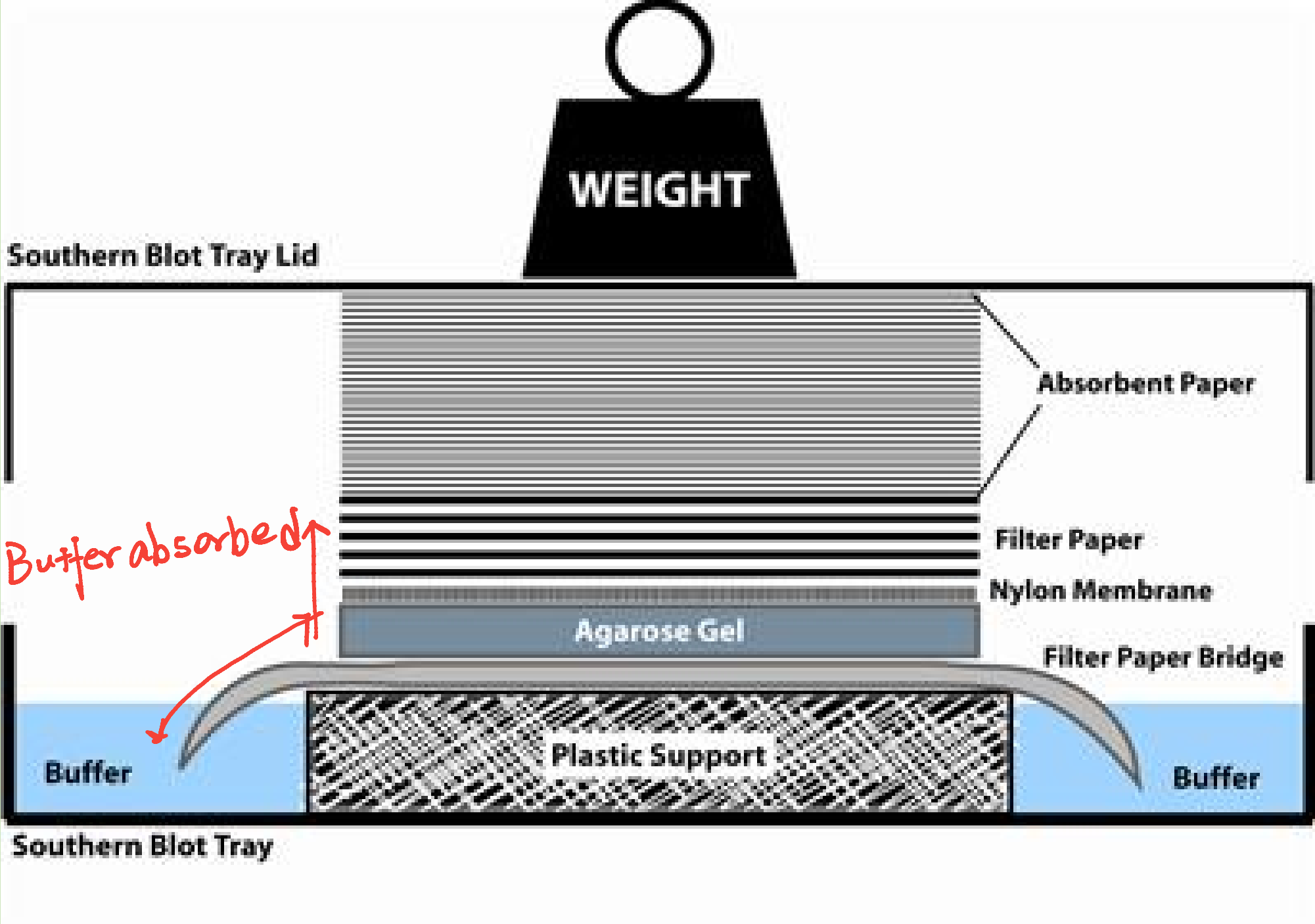
Denature of dsDNA: Alkaline denaturation
At high pH, then, the solution is rich in hydroxide ions, and these negatively-charged ions can pull hydrogen ions off of molecules like the base pairs in DNA. This process disrupts the hydrogen bonding that holds the two DNA strands together, causing them to separate.
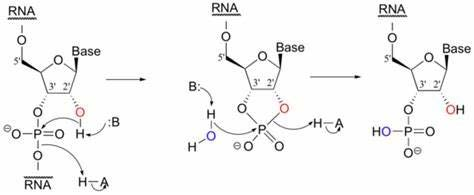
Unlike RNA, DNA lacks a hydroxyl group on the 2’ position in each sugar group. This difference makes DNA much more stable in alkaline solution.
In RNA, the hydroxyl group on the 2’ position can give up a hydrogen ion to the solution at high pH, creating a highly reactive alkoxide ion that attacks the phosphate group holding two neighboring nucleotides together.
- DNA does not suffer from this defect and thus enjoys remarkable stability at high pH.
2. Northern Blot

3. Western Blot

- Load and separate protein samples on SDS-PAGE
- Electrophoretically transfer fractionated proteins onto PVDF membrane
- Block the membrane with neutral protein (BSA or milk casein)
- Incubate the membrane with primary antibody specific to target protein
- Incubate the membrane with Hrp-labeled secondary antibody specific to primary antibody
- Incubate the blot with chemiluminescent HRP substrate and expose to film
4. Reporter Enzymes in Blotting- HRP vs AP
The most common reporter enzymes are horseradish peroxidase (HRP) or alkaline phosphatase (AP). Chemiluminescent substrates for both HRP and AP are very sensitive and can allow picogram to femtogram level detection.
- The maximum signal generated from HRP substrates is very fast, often within 5 minutes.
- The signal from AP substrates, however, gradually increases over time with a signal plateau around 60 minutes. The signal from AP substrates is very stable and can last up to several days, which can be useful if multiple exposures are needed
Generally, dilution factors for the AP 2° antibodies can be greater than with HRP antibodies, thus saving reagents. The disadvantages of either HRP or AP system are interferences, such as azides for HRP or phosphate buffers for AP. There is also the possibility that endogenous phosphatases will react with AP substrates.
Type of Filters in blotting techniques
Nitrocellulose Membrane (NC) - For the detection of nucleic acid and Protein.
- DNA/RNA covalently bound to membrane.
Positively charged Nylon Membrane – For DNA/RNA only
PVDF Membrane – For Protein only, a pre-treatment with methanol is required
Principles of molecular hybridization
Hybridization involves annealing between two strand of DNA and or RNA with partially or fully complementary sequence.
- T
m(Melting Curve)- Tm, Heating the DNA solution above a characteristic temperature can separate the two strands of a double helix. That temperature is called the melting temperature
- T
m50: 50% DNA become single stranded - Factors affect Tm would determine the stringency of hybridization
- High stringency means a condition only allows perfect match between two piece of ssDNA. (高严格度)
- Stringency of hybridization (Influenced by the following reasons)
- GC content
- GC content, Because a G–C base pair has three hydrogen bonds and an A–T pair only has two, nucleic acid double helices with a high G+C content have a higher Tm than do those with a greater proportion of A+T.
- Salt concentration
- Salt: High Salt concentration reduce stringency, Low salt concentration enhance stringency.
- A high concentration of salt (such as NaCl) raises the Tm of a DNA duplex, because the positive Na+ ions shield the negative charges on the phosphodiester backbone from repelling each other
- High salt decrease the stringency & Low salt increase the stringency – of hybridization
- Oppose to the protein-protein interaction
- Solvent
- Organic Solvent: DMSO, DMF, Formamide etc, certain organic solvents can cause the negative charges on the phosphates to repel more strongly, thus reducing the Tm.
- Create hydrophobic environment $\rarr$ de-stable hydrogen bonds
- GC content
Blotting techniques have been widely employed for more than 30 years and have provided the foundation of our understanding of molecular biology.
- However, these techniques have been largely—and in some cases completely— usurped by new technologies .especially the next generation sequencing techniques
Southern blots have been replaced by multiple techniques.
- Real-time PCR: boasts incredible sensitivity; theoretically, this method is able to detect even a single copy of the target sequence and compare relative copy numbers across samples rapidly and reliably, with little technical expertise required
- Fluorescent in situ hybridization (FISH) allows detection of specific sequences within a tissue sample with high sensitivity and precise localization
Northern blots have given way to reverse-transcription real-time PCR, again a more sensitive and more user-friendly technique.
三、Labeling methods of Nucleic Acid
Radioactive and non-radioactive labelling
- Radioactive labelling: Unbeatable sensitivity, Common radioactive isotope used in in molecular biology: ^32^P, ^3^H, ^125^I, ^35^S. ^14^C.
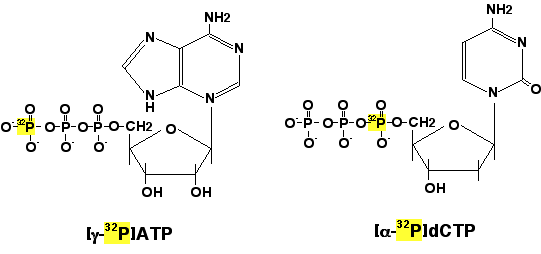
- Gamma-^32^P ATP used in the End Labelling
- Alpha- ^32^P dNTP can be used in any elongation reaction mediated by polymerase

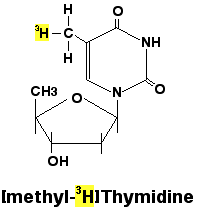
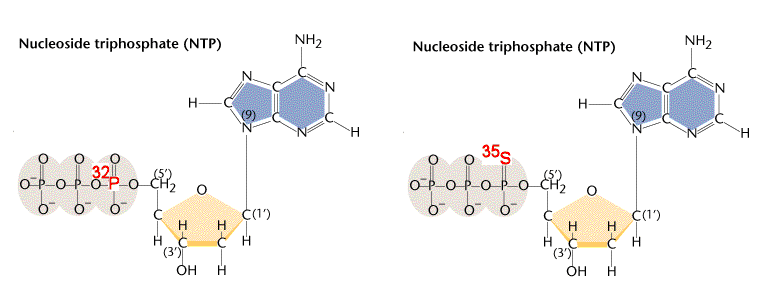
Common labeling methods of Nucleic Acid
- End labelling 末端标记
- Nick translation 切口平移
- Primer extension 引物延伸
- PCR labeling
1. 5’ End Labeling
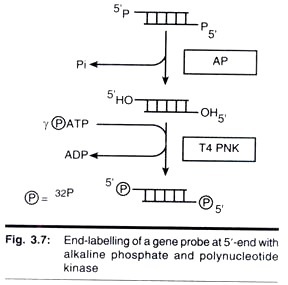
- AP: Alkaline phosphatase 碱性磷酸酶
- PNK: Polynucleotide Kinase 多聚核苷酸激酶
- Highly efficient, accurate
- Specific activity is relatively low (因为只标记了末端,中间部分没有标记)
2. Nick Translation
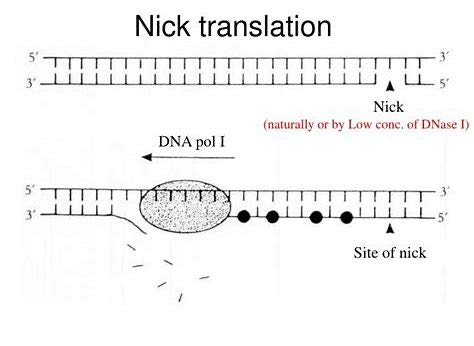
Nick: Low concentration of DNase I breaks the DNA strand, remove one nucleotide and leaves 5’ P and 3’ OH.
- E.coli DNA polymerase I has 5’ to 3’ exonuclease activity Polymerase activity
- One strand is degraded and replaced by an newly synthesized strand
Produce relatively short DNA(Radioactively labelled).
- Need lots of templates (no amplification)
- Accurate – amplification free
- Label both strand 可以实现对两条链都进行标记
3. Primer extension

- DNA is denatured to give single-stranded molecules.
- An oligonucleotide primer is then added to give a short double-stranded region with a free 3-OH group.
- The Klenow fragment of DNA polymerase I can then synthesize a copy of the template strand from the primer, incorporating [α-^32^P]DNTP(solid circles)to produce a labelled molecule with a very high specific activity
Primer;
- Klenow fragment
- Strand-specific
- High specific activity
4. Random prime Labeling
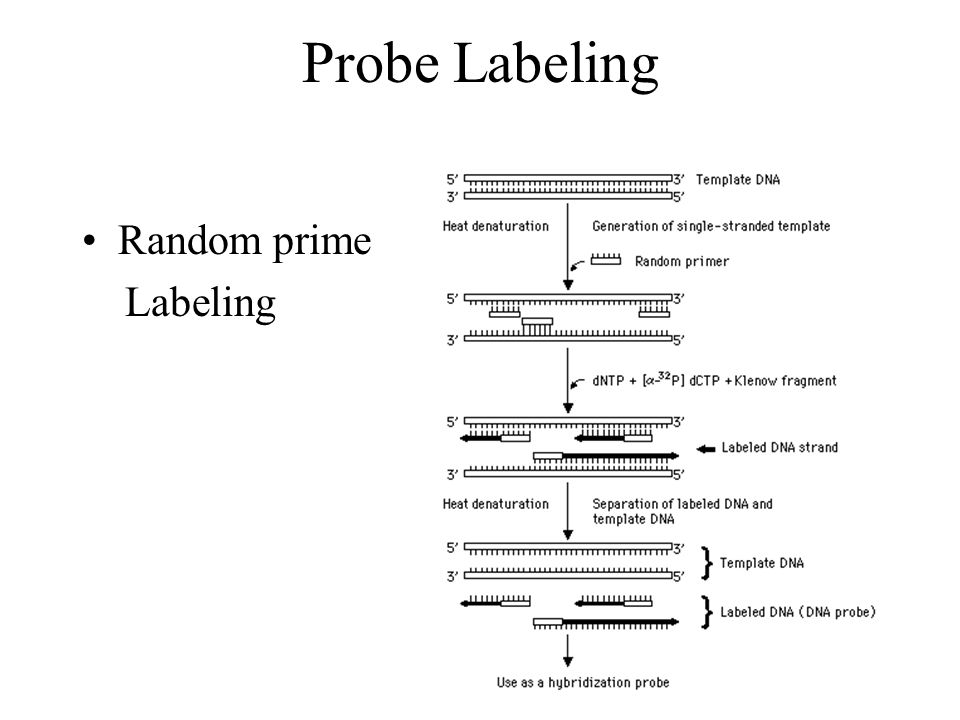
在DNA单链条件下,加入随机的(并不局限于特定序列的)引物,进行扩增。最终实现标记。
Non-radioactive labeling
- Digoxigenin-11-dUTP Anti-DIG-AP
- Biotin-11-dUTP Streptavidin-HRP
- Cy3/Cy5 – UTP Excitation-Emission
- 这些方法适用于copy数高,表达量高的基因
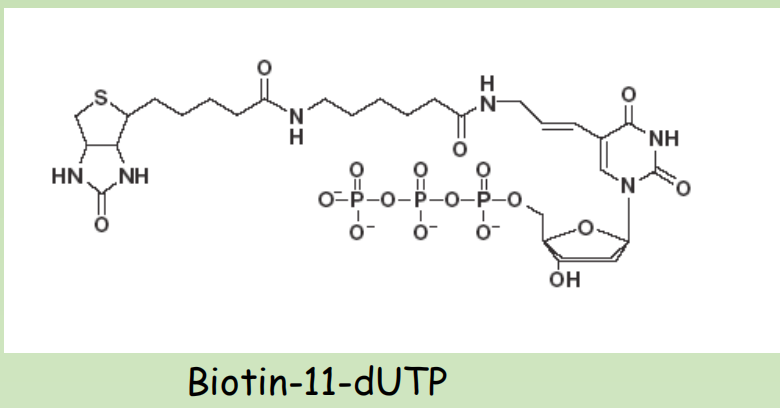
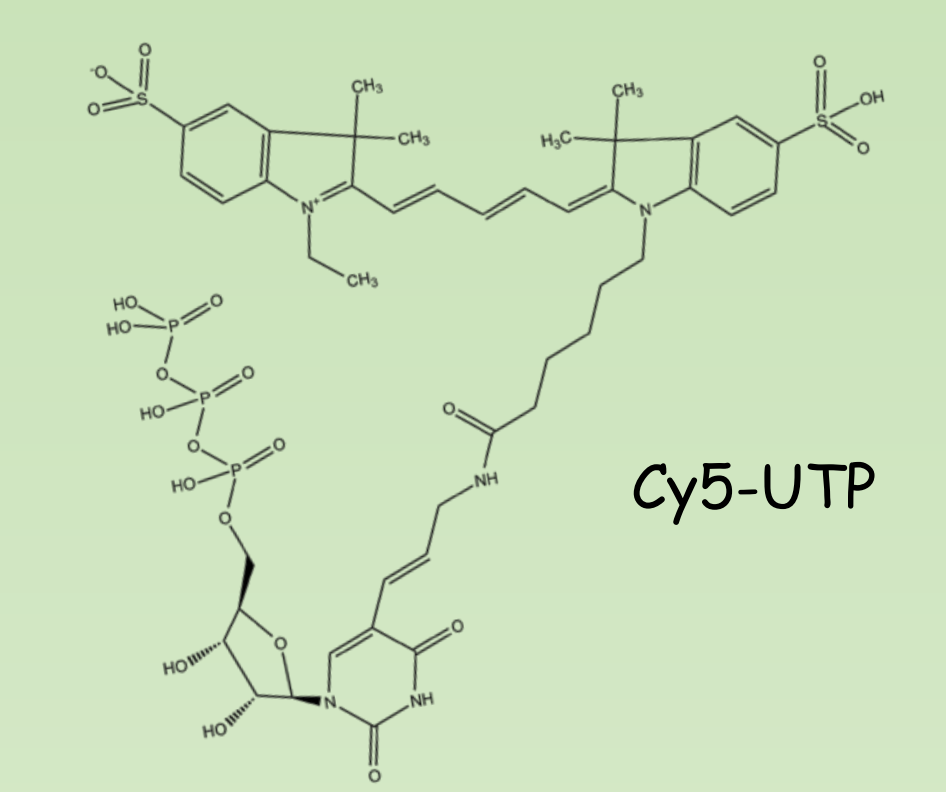
Detection of signal after hybridization

- ^32^P mainly emit beta particles
- This can be detected by X-ray film or The phosphor screen
- Phosphor screen can stably absorb energy and then release it upon laser stimulation
- Fluorescence is often being detected if nonradioactive labelling method is used
Summary
Handling and Quantification of DNA/RNA
- Absorbance vs Fluorescence based – OD260/280, OD260/230 Electrophoresis (Pros and Cons of different matrix)
Nucleic acids hybridization – different blotting techniques
- Southern/Northern blot/Western, Principle of hybridization (Idea of stringency), Type of Membrane, Alkaline Denaturation/Hydrolysis
Labeling methods of Nucleic Acid
- Radioactive VS Non-radioactive – common isotopes – common labeling methods –Signal detections.