L3 Isolation of DNA and RNAs
一、The Basic Lab Equipment
Getting to know common lab equipment
- Centrifuge
- Fridge and Freezer
- Vortex
- Water Bath
- PCR cycler, Thermo cycler
- Gel tank, Gel tray and comb
- Electrophoresis Power Supply
- Pipette and Pipette Tip
- Rotator
- Petri-dish
- Incubator
- Dry bath
- Sonicator
- Shaker
- Tube rack
- Filter
- Eppendorf Tube
- Falcon Tube
- Nanodrop
- Rotor, Fixed Angle,
- Swinging Bucket
二、Isolation of DNA and RNAs
General Considerations
The considerations behind a DNA extraction procedure:
- Maximize DNA recovery
- Remove inhibitors (which may interfere with down-stream applications)
- Remove or inhibit nucleases
- Maximize the quality of DNA
1. The Amount of DNA
The amount of DNA we can get
- The PCR reactions call for on average 1 ng of DNA (single or double stranded)
- Many of the commercially available kits are sensitive below 1 ng of DNA(100-250 pg)
- A southern blot will need ~ 1 to 10 ug of DNA.
The amount of DNA we can recover (The DNA in different subjects)
- A haploid human genome contains 3.2*10^9^ nucleotides
- Molecular weight of dsDNA=(No. of Nucleotides x 607.4) + 157.9, MW of human genome = 1.94 * 10^12^ g/mol
- 6.02×10²³ molecule per mol
- A haploid human cell contains (1.94 * 10^12^ / 6.02×10²³)*10^12^ = 3.2 pg of DNA
- The average WBC of an adult is 5 - 10 * 10^3^ Cells of 1μl Blood, equals to 60ng DNA
However, the yield of DNA is not a big problem for we have PCR.
2. The Major Steps
Cell Lysis
- Add lysis buffer to lyse the cell, protect the DNA/RNA from degradation
Separate the nucleic acids from Proteins/saccharide
- Phenol-chloroform extraction / binding of DNA/RNA to column
Obtaining DNA/RNA which is dissolved in small amount of solution.
- Elution, precipitation
Cell Lysis:
Purpose: Break the cell and release the cell content.
Some important reagents’ function:
EDTA: EDTA buffer inhibits some metal-dependent enzymes such as DNAseI.
Tris-Cl: The buffering reagent, maintain the overall pH.
Salt: (Nacl and Kcl) Provide salt and proper osmotic pressure.
SDS: The ionic detergent. Disrupt the cell membrane and denature the protein. In order to promote the release of cell content.
Phenol-Chloroform Extraction:
- Function: Separate the DNA molecules from other unnecessary substances (protein, carbohydrates, lipids, etc.)
- Some important reagents’ function:
- Phenol: Insolubilize protein and denature proteins.
- Chloroform: Denature lipids and separate the organic and aqueous phase.
- Isoamyl alcohol (Alternative): Prevent foaming in the interphase and emusification, therefore we can get clear sample.
DNA Precipitation:
- Function: Collect the DNA molecule for the future experiments.
- Some important reagents’ function:
- Salts: Naturalize the negative charge in DNA molecule and therefore make it less hydrophilic.
- 70% Ethanol: Wash away the unnecessary reagents used in previous step, such as salts, organic reagents, etc.
- TE buffer: Dissolves DNA and then protect it from degradation.
3. The Reagents Used in DNA/RNA Isolation
Cell Lysis Buffer
Cell lysis buffer – typical lysis buffer
- Buffering reagent
- Tris, HEPES, MES, PIPES. 10-100mM
- Detergent Nonionic detergent, Ionic detergent
- Nonionic detergent: Triton X-100, NP40, Tween 20
- Ionic detergent: SDS, CTAB, Deoxycholate, Concentration: 0.5%-2%
- Function: Disrupt membrane and denature proteins.
- Salt
- NaCl, KCl
- physiological concentration – 100mM to 1M
- Provide salt and osmotic pressure.
- NaCl, KCl
- Chelating reagent
- EDTA
- EDTA buffer inhibits metal-dependent enzymes such as DNase I
- Antioxidant
- 2-Mercaptoethanol, DTT
- providing reducing environment, or to reduce disulfide bonds
- 2-Mercaptoethanol, DTT
Definition of nonionic detergent
- Any of a class of synthetic detergents (as long-chain ether derivatives or esters of alcohols or phenols) that are neither anionic nor cationic but produce electrically neutral colloidal particles in solution

Definition of ionic detergent
- Ionic detergents have a hydrophilic head group that is charged and can be either negatively (anionic) or positively (cationic) charged. Ionic detergents are used for the complete disruption of cellular structures and denaturation of proteins for separation during gel electrophoresis. Ionic detergents bind with protein molecules, masking their native charge and rendering the protein molecules the overall charge of the ionic detergent.

Used in DNA/RNA Isolation
Separate the nucleic acids from Proteins/polysaccharide
- Organic Extraction (Phenol-Chloroform)
- Non-Organic (Proteinase K and Salting out)
- Column based method
The method utilized may be sample dependent, technique dependent, or analyst preference
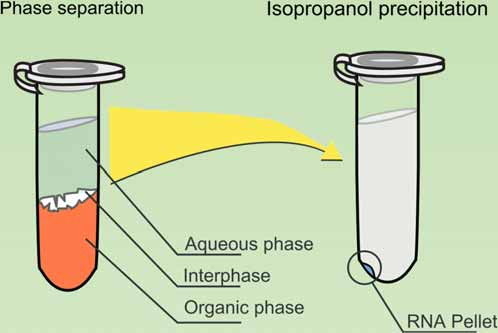
(1) Phenol-chloroform extraction
Deproteinization:
- Phenol can denature the proteins, which will then be precipitated at the interphase or trapped in the organic phase.

Phenol:
- DNA is insoluble in phenol because phenol is a non-polar solution
- Protein has both polar and non-polar group present in it because of the long chain of different amino acids. Different amino acids have different groups present on their side chain. Also, the folding of the protein into the secondary, tertiary and quaternary structure depends on the polarity of the amino acids.
- The bonds between amino acids are broken by the addition of phenol and protein get denatured
- Denature DNA and insolubilize protein.
Chloroform:
- chloroform allows proper separation of the organic phase and aqueous phase which keeps DNA protected into the aqueous phase. Chloroform denatures the lipid as well.
Isoamyl alcohol:
- In the phenol-chloroform DNA extraction method, Isoamyl alcohol helps in reducing foaming(防止起泡沫) between interphase. It prevents the emulsification of a solution.
- The liquid phase contains DNA and the organic phase contains lipid, proteins and other impurities. The precipitated protein denatured and coagulated between both these phases (interface). This will create the cloudy, whitishfoam between interphase. The anti-foaming agent, isoamyl alcohol stabilized the interphase by removing the foaming. This will increase the purity of DNA.
Used in DNA/RNA Extraction
Obtaining DNA/RNA which is dissolved in small amount of solution. → elution, precipitation
The overall principle of ethanol precipitation : elution, precipitation
- Nucleic acids are hydrophilic due to the negatively charged phosphate (PO3-) groups along the sugar phosphate backbone.
- Because of these charges, polar molecules, like DNA or RNA, can interact electrostatically with the water molecules, allowing them to easily dissolve in water.
Role of Salts:
- positively charged sodium ions neutralize the negative charge on the PO3- groups on the nucleic acids, making the molecule far less hydrophilic, and therefore much less soluble in water
- H
2O is very polar $\rarr$ salt is hard to meet DNA $\rarr$ ethanol
The role of the ethanol (70%):
- Water has a high dielectric constant, which makes it fairly difficult for the Na+ and PO3- to come together.
- Ethanol on the other hand has a much lower dielectric (非传导性的) constant, making it much easier for Na+ to interact with the PO3-, shield it’s charge and make the nucleic acid less hydrophilic, causing it to drop out of solution.
The role of temperature in ethanol precipitation:
- Incubation of the nucleic acid/salt/ethanol mixture at low temperatures (e.g. -20 or -80C) is commonly cited in protocols as necessary in protocols.
- However, according to (Molecular Cloning, A Laboratory Manual 2nd Edition), this is not required, as nucleic acids at concentrations as low as 20ng/mL will precipitate at 0-4C so incubation for 15-30 minutes on ice is sufficient
Glycogen:
- Glycogen is a polymer which is used to trap the nucleic acids (DNA, rRNA,RNA…). In ethanol, Glycogen is insoluble so it forms polymer structure and traps the nucleic acids and forms pellet with nucleic acids trapped during the centrifugation…
Choice of salt:
- Use Sodium acetate (0.3M final conc, pH 5.2) for routine DNA precipitations
- Use Sodium chloride (0,2M final conc) for DNA samples containing SDS since NaCl keeps SDS soluble in 70% ethanol so it won’t precipitate with the DNA.
- Use Ammonium acetate (2M final conc) for the removal of dNTPs, but do not use for preparation of DNA for T4 polynucleotide kinase reactions as ammonium ions inhibit the enzyme
- Use Lithium Chloride (0.8M final conc) for RNA. This is because 2.5-3 volumes of ethanol should be used for RNA precipitation and LiCl is more soluble in ethanol than NaAc so will not precipitate
- but beware – chloride ions will inhibit protein synthesis and DNA polymerase so LiCl is no good for RNA preps for in vitro translation or reverse transcription. In these cases, use NaAc.
4. Column based purification

- Step 1: Cell Lysis
- Step 2: Purification – Binding nucleic acids to the column
- Step 3: Washing
- Step 4: Dry Spin for ethanol-free DNA and RNA
- Step 5: The final frontier – Elution
The Cell Lysis
what’s the difference compared with phenol/chloroform method ?
- The key is chaotropic salts(促溶盐;离液盐;离序盐) - guanidine HCL, guanidine thiocyanate, urea, and lithium perchlorate.
- The term “chaotropic“ means chaos-forming, a term which in biochemistry usually refers to a compound’s ability to disrupt the regular hydrogen bond structures in water
- Hydrogen bonding profoundly affects the secondary structure of polymers such as DNA, RNA, and proteins
- Hydrogen bonding also affects how water-soluble a molecule is. With the addition of chaotropic ions to the nucleic acid, the ordered structure of water molecules of its hydrate shell is destroyed.
The Lysis Buffers:
- Lysis buffers contain a high concentration of chaotropic salts
- Chaotropes have two important roles in nucleic acid extraction
- Firstly, they destabilize hydrogen bonds, van der Waals forces and hydrophobic interactions, leading to destabilization of proteins, including nucleases.
- Secondly, they disrupt the association of nucleic acids with water, thereby providing optimal conditions for their transfer to silica.
One possible explanation for functions of salt:
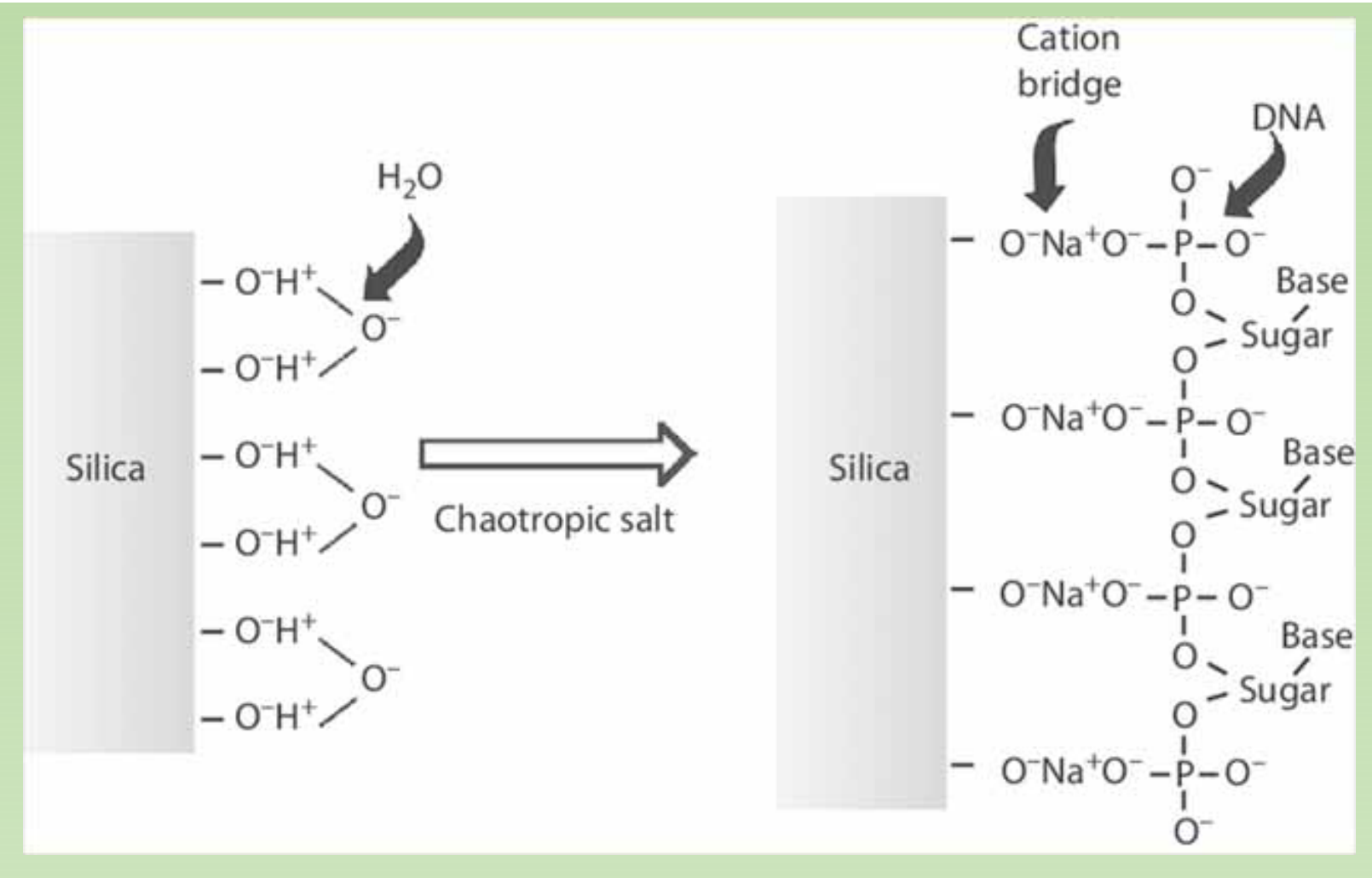
The exact mechanism how the silicon adsorption columns work is not quite clear. One idea is as follows. Chaotropic agent guandium HCl, which disrupt water structure, allow positively charged ions to form a salt bridge between the negatively charged silica and the negatively charged DNA backbone under high salt conditions.
Purification
Step 2: Purification – Binding nucleic acids to the column
- Addition of ethanol in the cleared lysate.
- The addition of alcohol (or sometimes isopropanol) will further enhance and influence the binding of nucleic acids to the silica.
- Too much ethanol will bring down degraded material and small species that will influence UV260 readings.
- Too little ethanol may impede washing of the salt from the membrane
Washing
Washing-To wash away protein and salt residues
- There are typically two wash steps.
- The first wash will often include a low concentration of chaotropic salts to remove residual proteins and pigments.
- This is always followed with an ethanol wash to remove the salts.
Dry Spin for ethanol
Dry Spin for ethanol-free DNA and RNA
Elution
For DNA extraction, 10 mM Tris at pH 8-9 is typically used. DNA is more stable at a slightly basic pH and will dissolve faster in a buffer than water. This is true even for DNA pellets. Water tends to have a lower pH of 4-5, and high molecular weight DNA may not completely rehydrate in the short time used for elution
DNA extraction vs. RNA extraction
For DNA – things to avoid: shearing force, DNase, Get rid of RNA if necessary.
- Avoid sharing force is to keep the target DNA fragment intact
For RNA – Inactivate endogenous RNase and avoid exogenous RNase, DNase digestion is almost always necessary for downstream applications.
Trizol RNA extraction

Trizol RNA extraction: An variety of Phenol/Chloroform method
Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156-9.
4 M guanidinium thiocyanate, 25 mM sodium citrate, 0.5% (wt/vol) N-laurosylsarcosine (Sarkosyl), 0.1 M 2-mercaptoethanol,50% Phenol, sodium acetate pH 4.0