L09 Transposons And Retrotransposons
Transposons VS. Retrotransposons
Eukaryotic DNA transposons move via a DNA intermediate, which is excised from the donor site.
Retrotransposons are first transcribed into an RNA molecule, which is then reverse-transcribed into a double-stranded DNA.
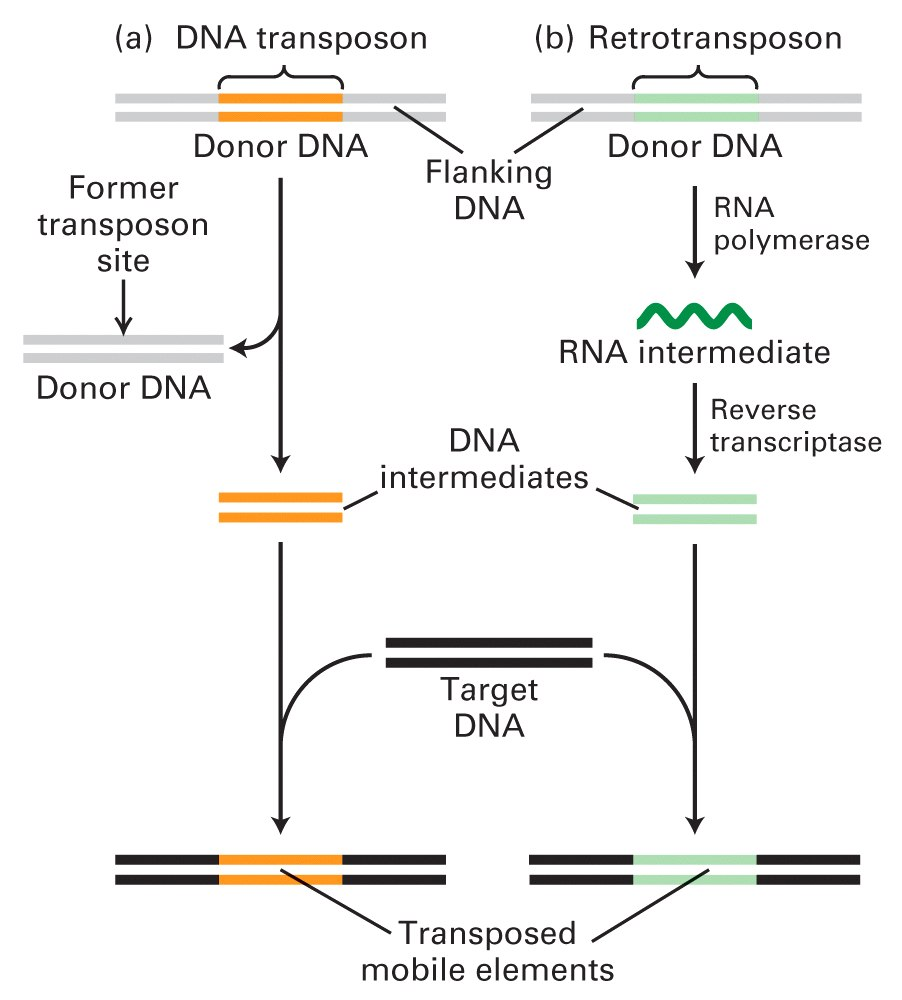
In both cases, the double-stranded DNA intermediate is integrated into the target site DNA to complete the movement. Thus, DNA transposons move by a cut-and-paste mechanisms, whereas retrotransposons(反转录转座子,RNA介导) move by a copy-and-paste mechanism.
Transposable Elements
一、Transposons
Transposable elements are pieces of DNA that can move into new locations in chromosomes = mobile DNA.
Since uncontrolled movement could wreak havoc on genomes, their transposition has evolved to be regulated.
Their transposition is useful for genetic analysis and gene isolation.
They are essentially opportunistic DNA sequences.
Transposons are found in bacteria, but play an important role in plant biology
Often have no phenotype themselves, but insertion has effect by knocking out genes.
DNA transposons mobilize via CUT & PASTE mechanism
Structure And Mechanism
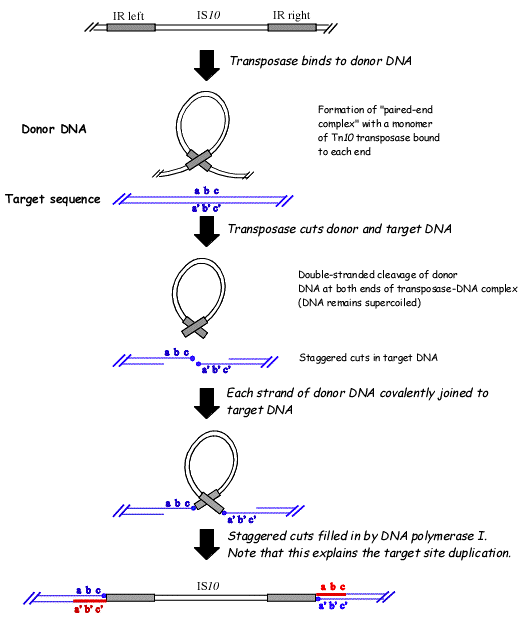
A common feature of transposable elements is the flanking of the element by short inverted repeat sequences. (通过短的反向重复序列将元件侧接)
A model for transposition suggests that the enzyme transposase (转座酶) recognizes these sequences, creates a stem/loop structure, and then excises the loop from that region of the genome. The excised loop can then be inserted into another region of the genome.
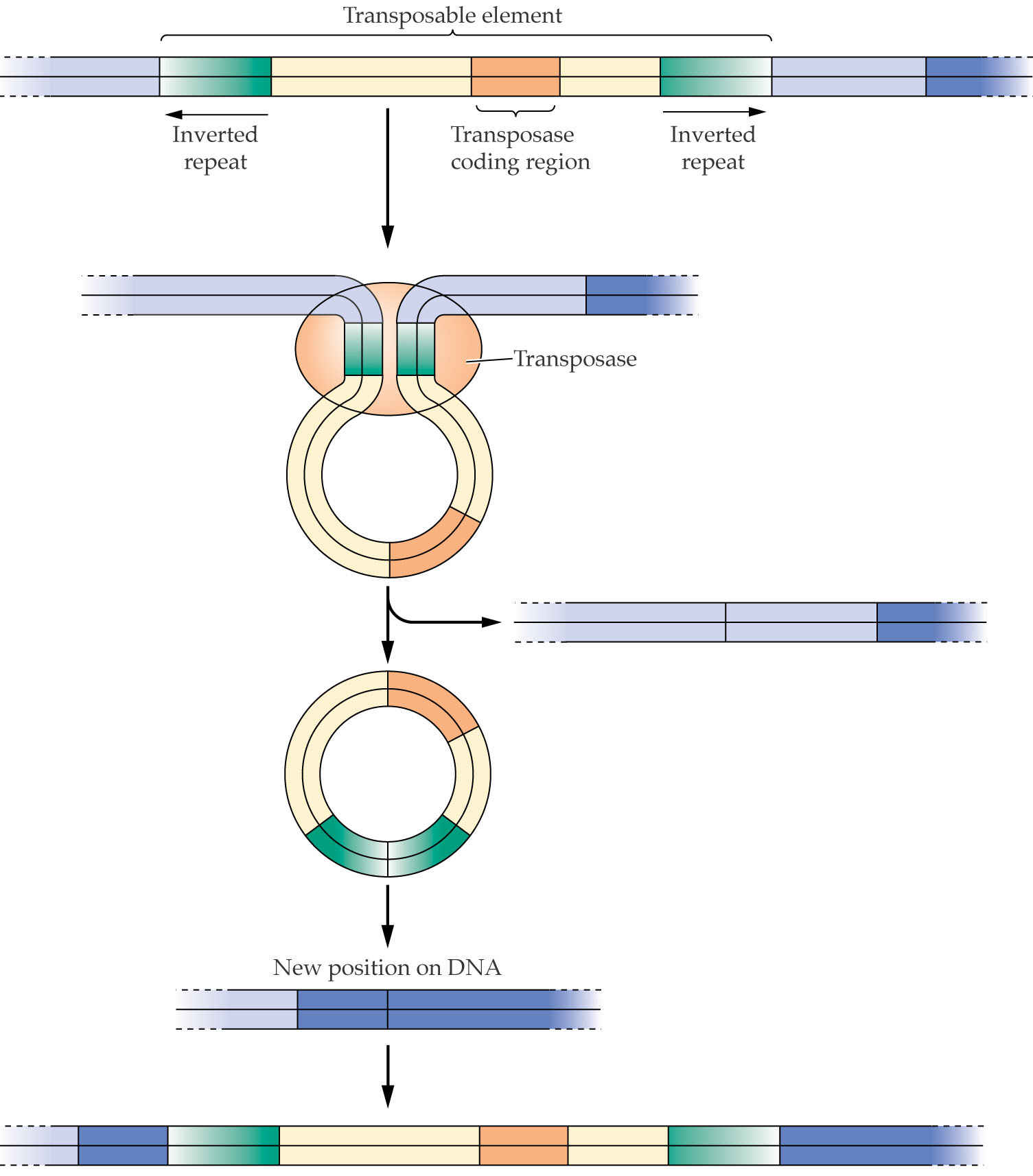
1. Transposase
DNA transposons mobilize via CUT & PASTE mechanism
The transposase cleaves the element, stays with the fragment, and reintegrates it at a new site.
This mechanism maintains a relatively low copy number in the genome.
Insertion of transposons causes duplication of the target site, due to the staggered(错列) cut at the target sequence, which is repaired by DNA polymerase.(这种错列将会由DNA聚合酶进行修理)
The Technological Use of Transposons
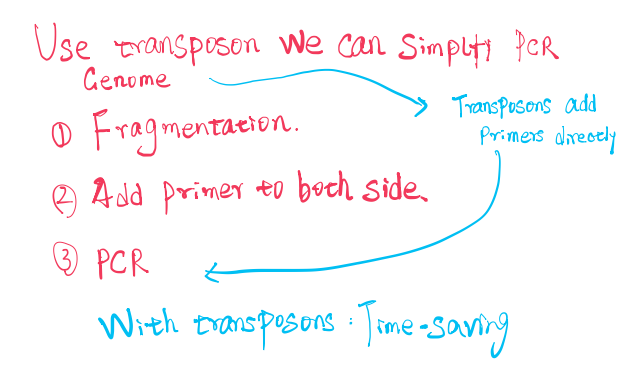
在PCR技术中,通过transposons直接插入primers,跳过fragmentation与Add primer的步骤,缩短实验时间
Transposons – Source of Mutation
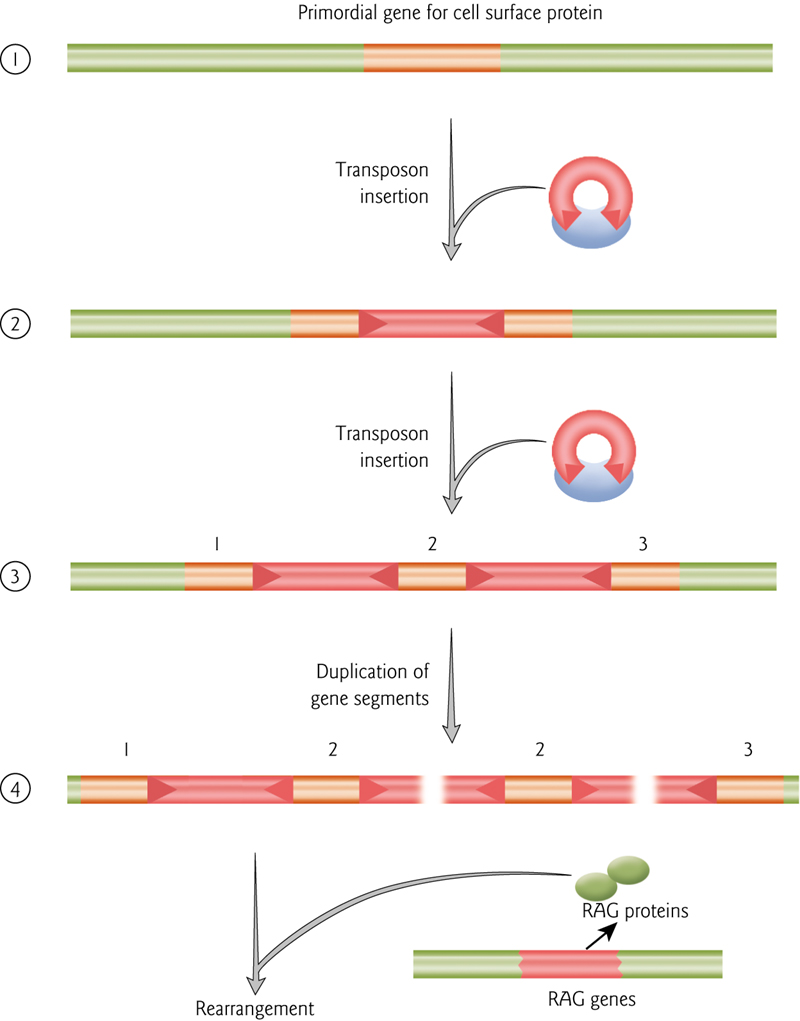
Amplification of DNA units and flanking transposon ends could create multiple transposase binding sites.
(DNA单元和转座子末端的扩增可能会产生多个转座酶结合位点。)
These could catalyze genomic rearrangements.
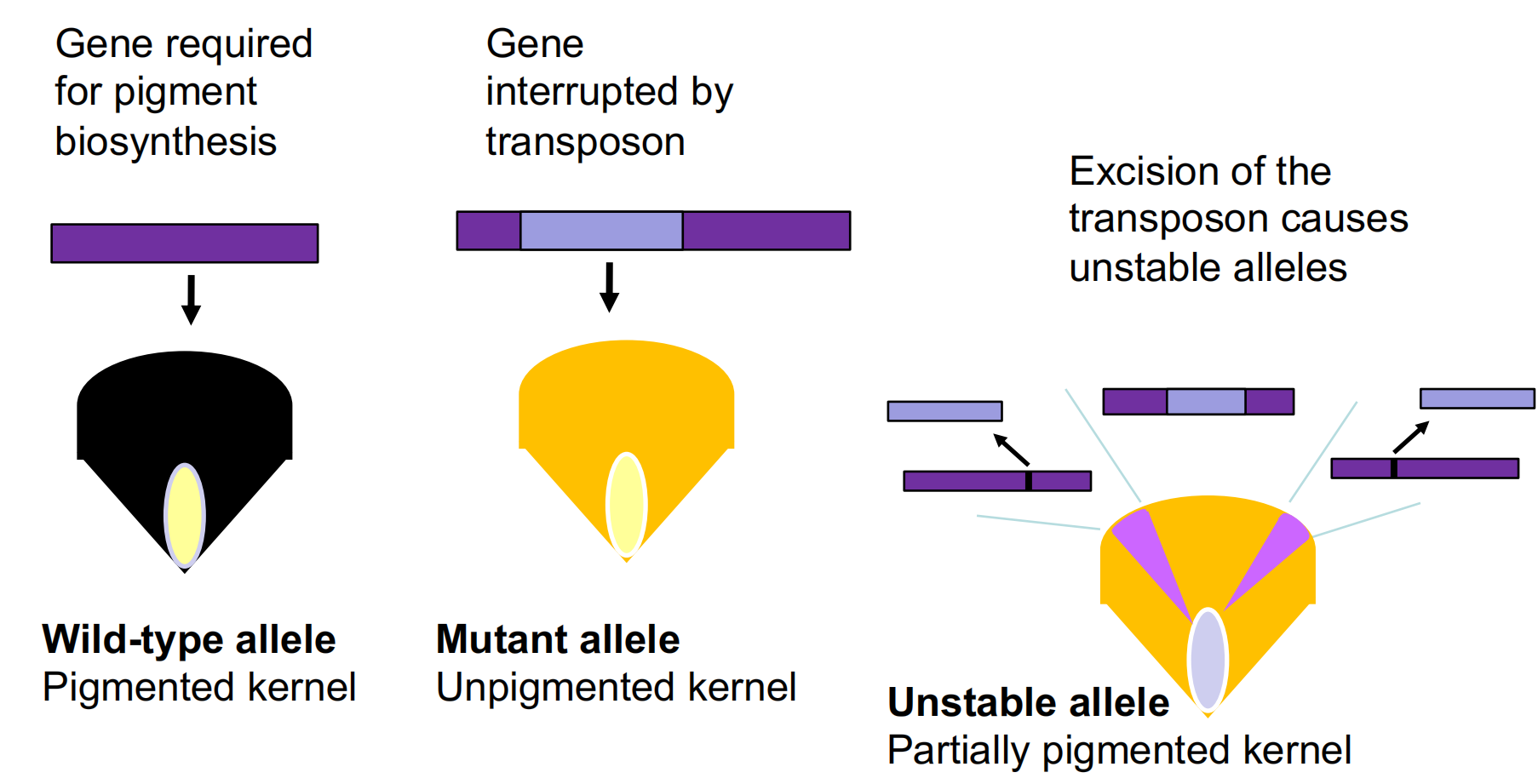
Transposons can cause inactive or unstable alleles.
Naturally occurring transposons are a source of genetic variation
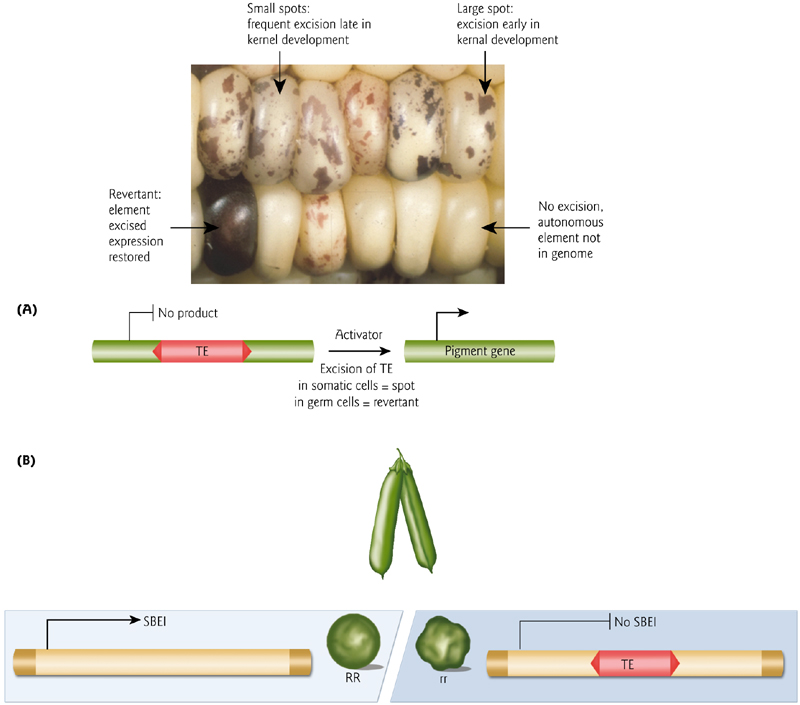
Transposons Excision Can Alter Gene Expressions
1. In Maize 玉米
Maize Ac/Ds Transposons
Transposable elements may be autonomous or non-autonomous.
Maize transposable elements of the “Ac” (Activator) type transpose regardless of the genetic background they are in. They belong to the class of autonomous elements.
Maize transposable elements of the Ds (Dissociation) type do not transpose unless one or more copies of Ac are also present in the genome. Therefore, Ds is a non-autonomous element.
Barbara McClintock’s Study:
- Ac: Can Transport by itself
- Autonomous elements encode a factor that can act in “trans” to move co-resident non-autonomous elements.
- Ds: Cannot transport by itself
- Non-autonomous elements do not encode a “cis-acting” factor (probably a transposase) needed for transposition.
- Non-autonomous elements may derive from autonomous elements by deletion of internal segments, thus inactivating their ability to program transposition.
- Alternatively, non-autonomous elements may arise when two sequences arise in proximity by mutation to be sufficiently similar to the element’s inverted repeat termini.
Ds (Dissociation) factor causes tendency toward chromosome breakage, but only if the individual also has Ac (Activator).
Ds could not be mapped – it seemed to occur in different places in the genome in different lines, and could move to a new place in different progeny; Barbara McClintock called this transposition.
Ds can cause unstable mutations, that frequently revert back to wild-type, but only if Ac is present.
Ds is a transposable element; it is non-autonomous, because it requires Ac to be present.
Ac encodes the transposase, the enzyme that catalyzes transposition.
Autonomous or Non-Autonomous
Transposable elements may be autonomous or non-autonomous.
Interpretations
Non-autonomous elements do not encode a “cis-acting” factor (probably a transposase) needed for transposition.
Autonomous elements encode a factor that can act in “trans” to move co-resident non-autonomous elements.
Non-autonomous elements may derive from autonomous elements by deletion of internal segments, thus inactivating their ability to program transposition.
Alternatively, non-autonomous elements may arise when two sequences arise in proximity by mutation to be sufficiently similar to the element’s inverted repeat termini.
Maize Transposons Acts In Arabidopsis.
The maize Ac element also functions in Arabidopsis.
Plants were transformed with a streptomycin-resistance gene interrupted by an Ac element and cultured to regenerate plantlets. So when this gene is interrupted, the plants are white due to loss of chlorophyll caused by streptomycin.
Excision of Ac from some cells yields streptomycin-resistant green sectors (i.e., restores chloroplast function) in a streptomycin-sensitive (white) background, shown in (A) and (B). This indicates that the Ac element has moved out of the streptomycin gene – and it is happening randomly in normal tissues.
2. Cause Gene Unstable or Inactive
3. Influence Neighboring Genes
“epigenetic” variation can arise due to quantitative effects of silencing on transposons that happen to be associated with genes due solely to their proximity(接近,附近) – which is a random effect.
In this example, a transposon in the promoter can cause flower color variation.
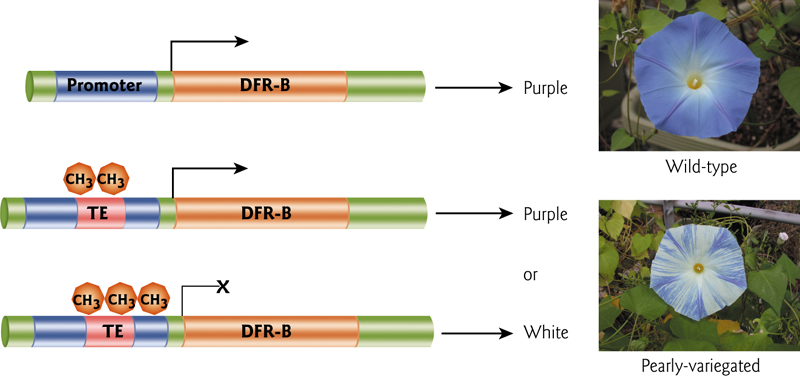
Same situation in mice – coat color is regulated by the methylation status of a retrotransposon in a “pseudoexon”. This element has a cryptic promoter that causes constitutive expression of the gene.
二、Retrotransposons
Retrotransposons mobilize via a COPY & PASTE mechanism
Reverse transcriptase makes DNA from RNA produced from the retroelement.
This can permit substantial increases in the total copy number of the element.
Most plant genomes expand via exactly this mechanism.
Retrotransposon Movement
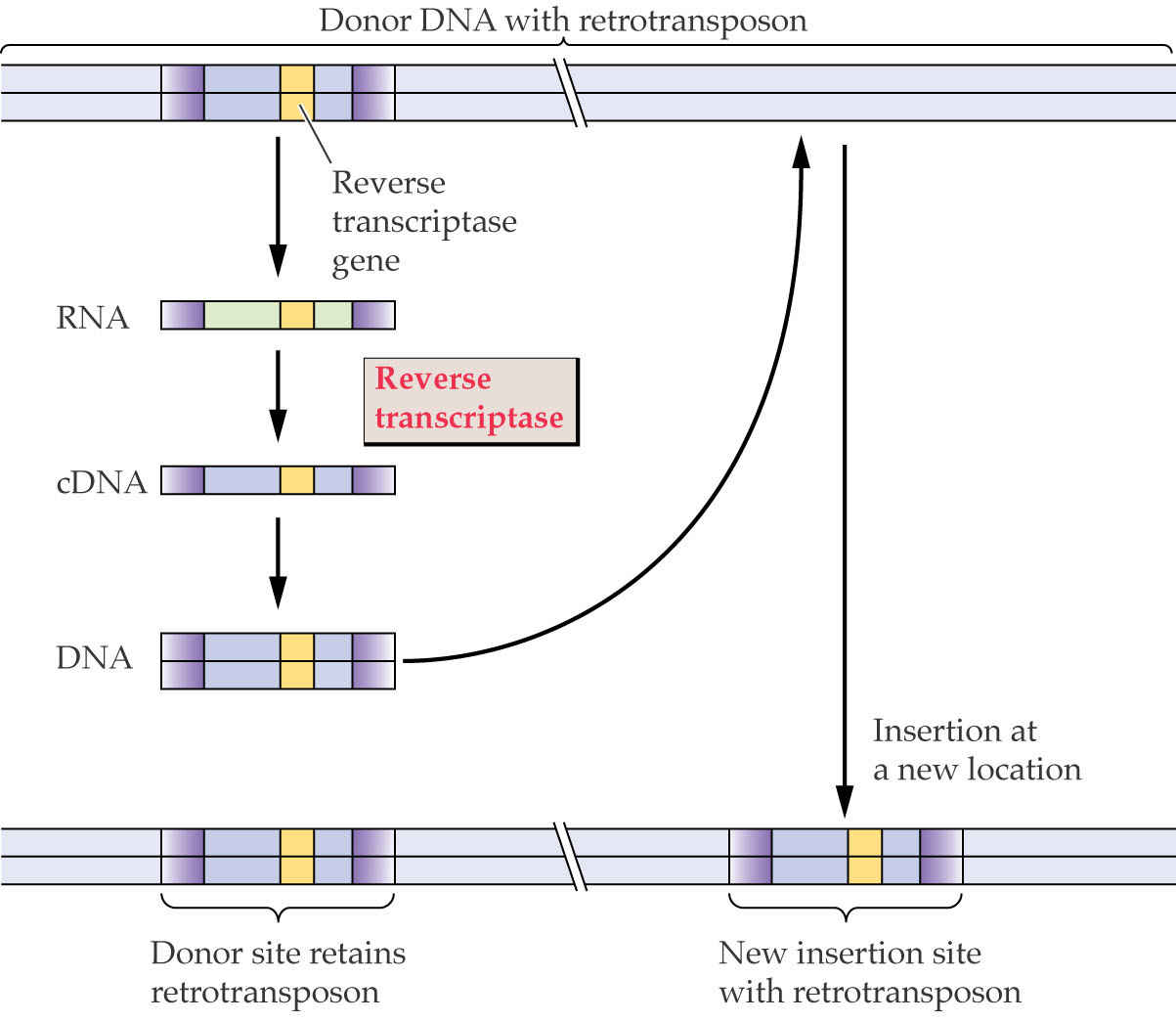
Retrotransposons transpose by a different means of transferring a section of DNA from one point to another compared to transposons. In this case, the element to be transposed is copied in place, through creation of an RNA intermediate, after which the duplicate copy is inserted into a new location.
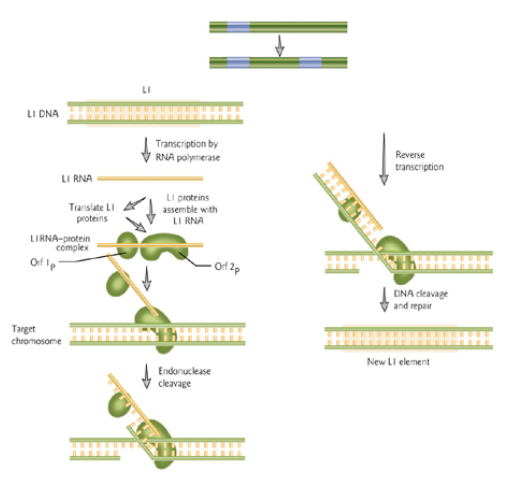
Overview of Retrotransposons
1. Phylogeny
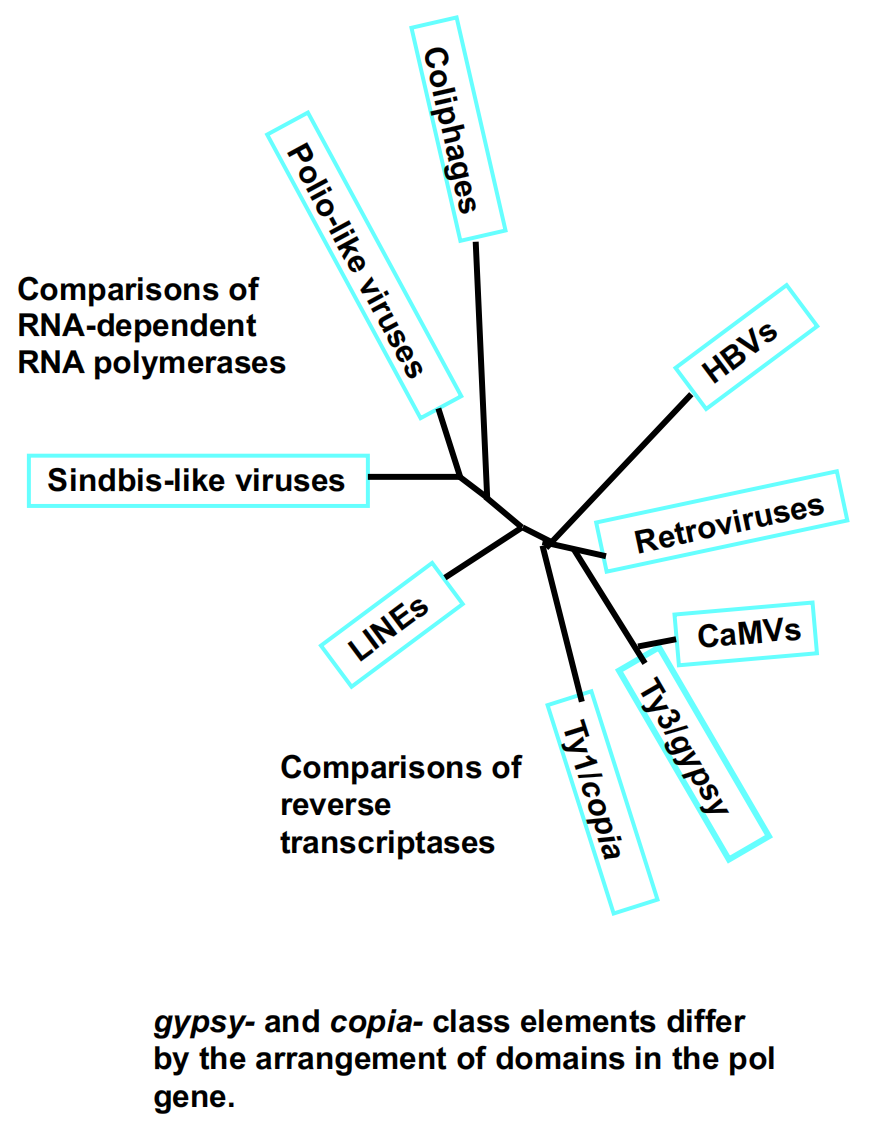
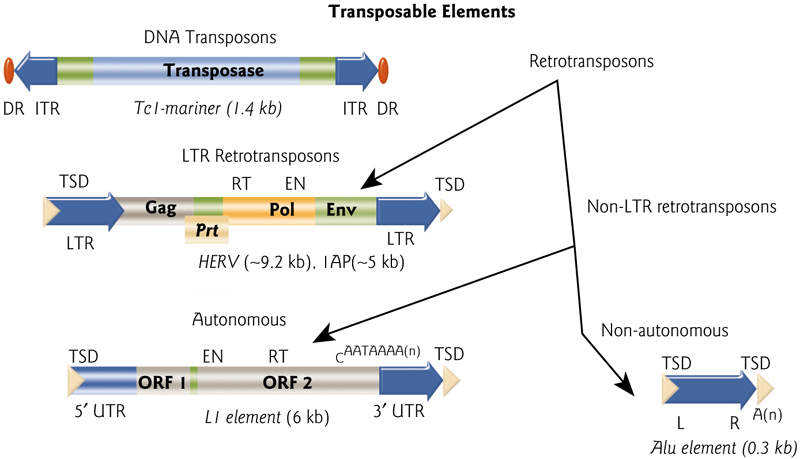
LTR Retrotransposons
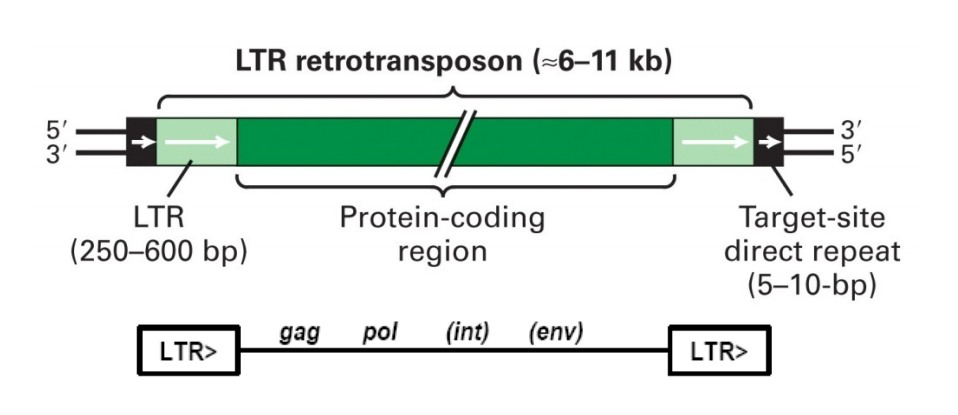
LTR retrotransposons are similar in structure and action to retroviruses, but never leave cell.
LTR = Long term repeat
Transcription starts within 5’ LTR, goes into 3’ LTR. The element may include genes that encode these proteins:
protease (gag)
reverse transcriptase (pol)
integrase (int)
viral coat protein (env)
Eukaryotic retrotransposons move via an RNA intermediate:
- The Transposase (TE) is transcribed by RNA polymerase
- A DNA copy is made from the RNA by the enzyme reverse transcriptase (RT)
- The DNA copy gets integrated into a new site in the genome by an integrase.
RT is encoded by the retrotransposon, not the host genome. The polymerase (“pol”)-like open reading frame is probably the gene for a reverse transcriptase that transcribes processed RNA into DNA.
Retrotransposons (LINE and SINE Transposons)
Non-viral retrotransposons do not have LTRs; most common TE in human genome (most are inactive and cannot move)
1. LINE
LINE = long interspersed element
Nick genomic DNA, and use DNA end to prime RT, which is often incomplete autonomous elements have both genes and promoter
Example: human L1, present in 100,000 copies per genome
2. SINE
SINE = short interspersed element
promoter derived from tRNA or another RNA gene
- no genes encoded; uses LINE machinery to move, therefore non-autonomous
Example: human Alu element, present in >106 copies (5% of our DNA)
In plants, similar elements called MITEs – 100 to 600 bp, form secondary structures.
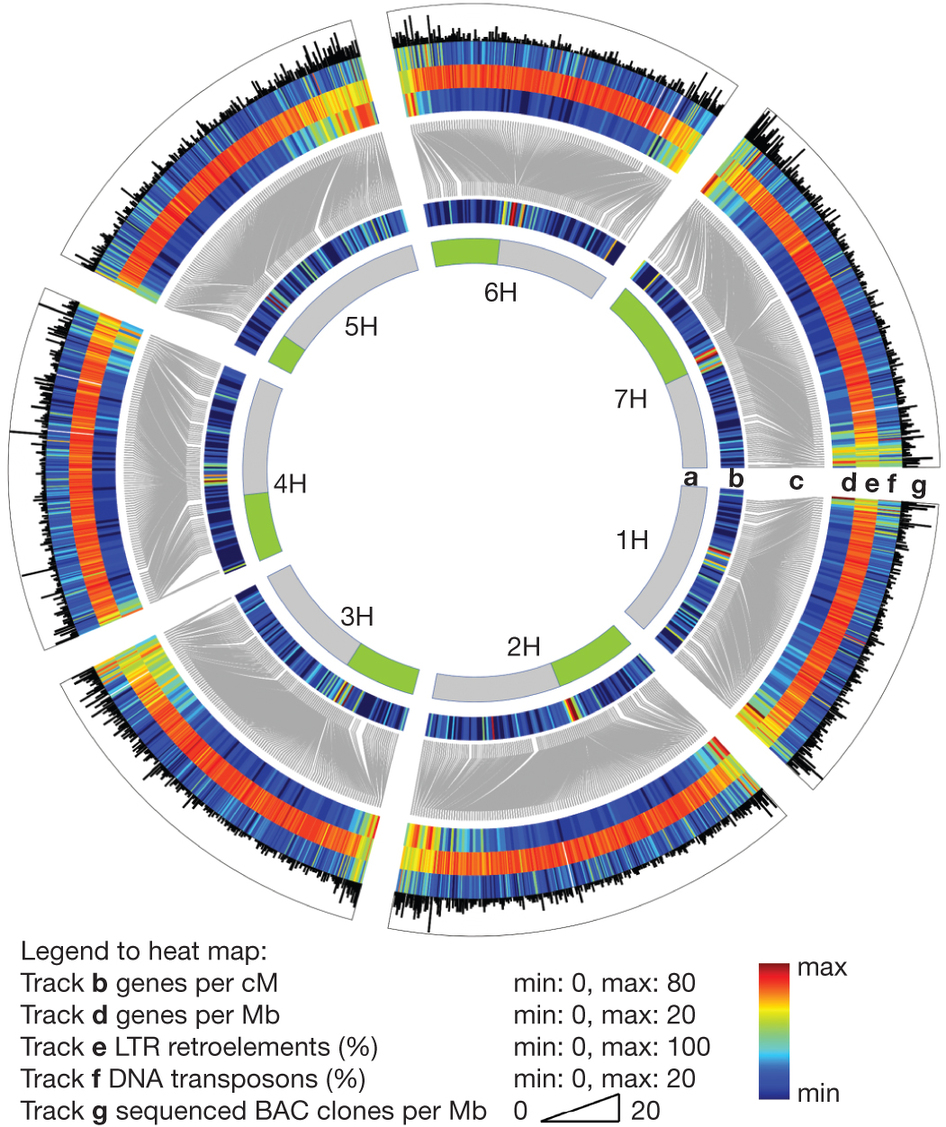
三、Nucleotide Sequence Content of Genome
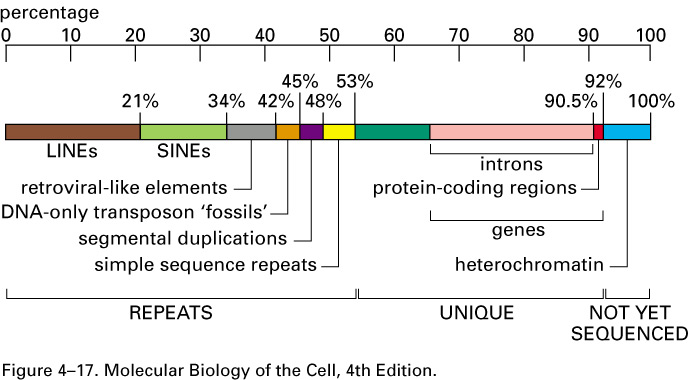
Human Genome
Representation of nucleotide sequence content of human genome
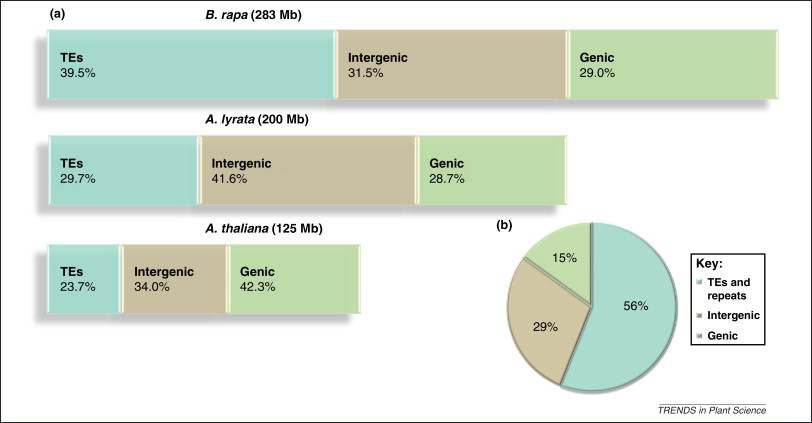
Plants Genome Size Are Different
Classical measurements of DNA content showed that plants vary considerably in genome size. Even among closely related species like grasses.
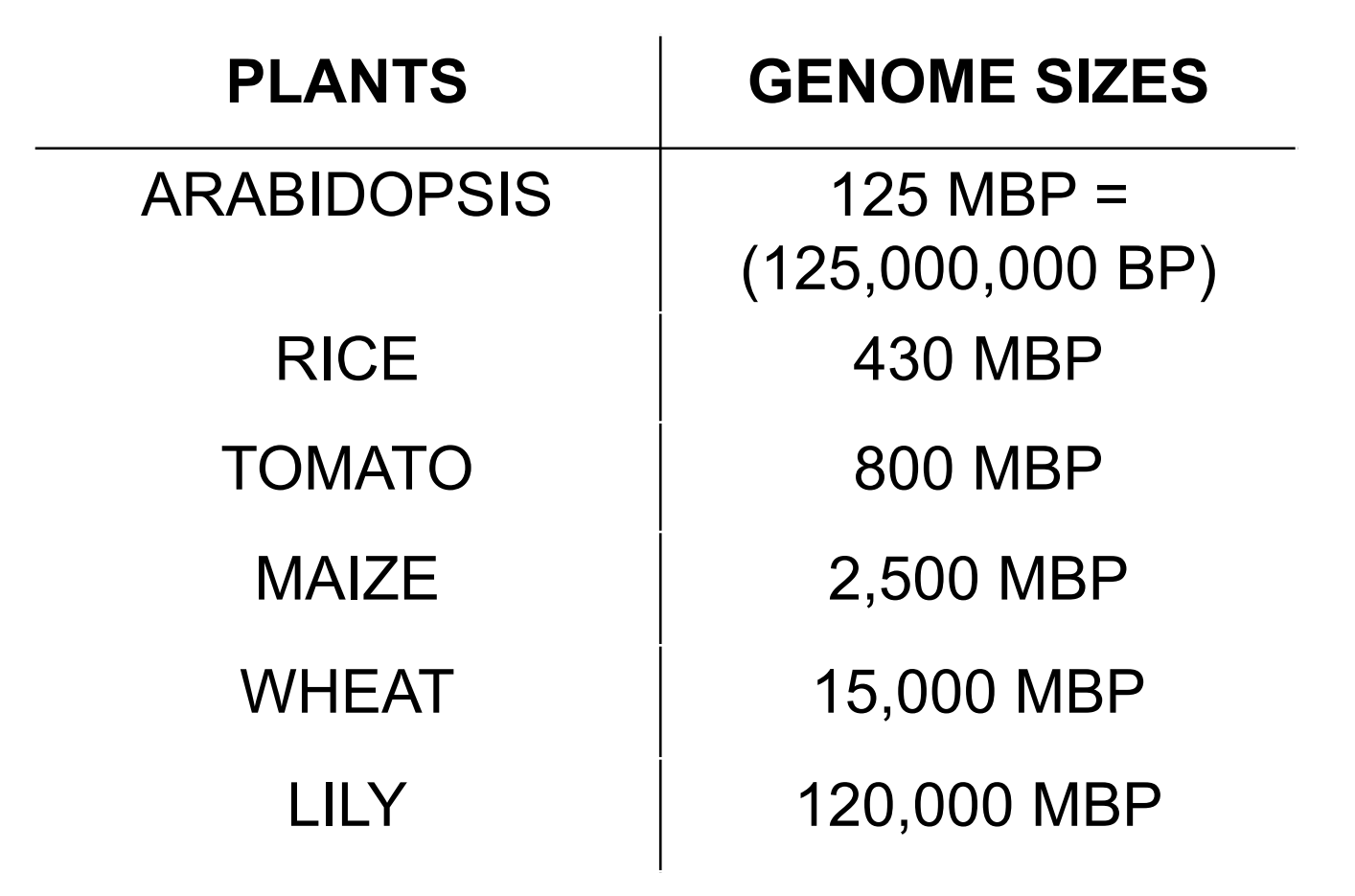
1. The Maize Genome

2. Arabidopsis
3. Barley 大麦