L10 Replication of DNA
一、Overview of DNA Replication
DNA replication goal ‐ make an exact copy of DNA
Must occur
- Very quickly – millions to billons of bases in minutes to hours
- Very accurately – few to no mutations (could be deleterious to daughter cells)
- At appropriate time in the life of the cell
Proposed Models of DNA Replication
1. Conservative model 全保留复制模型
Both parental strands stay together after DNA replication
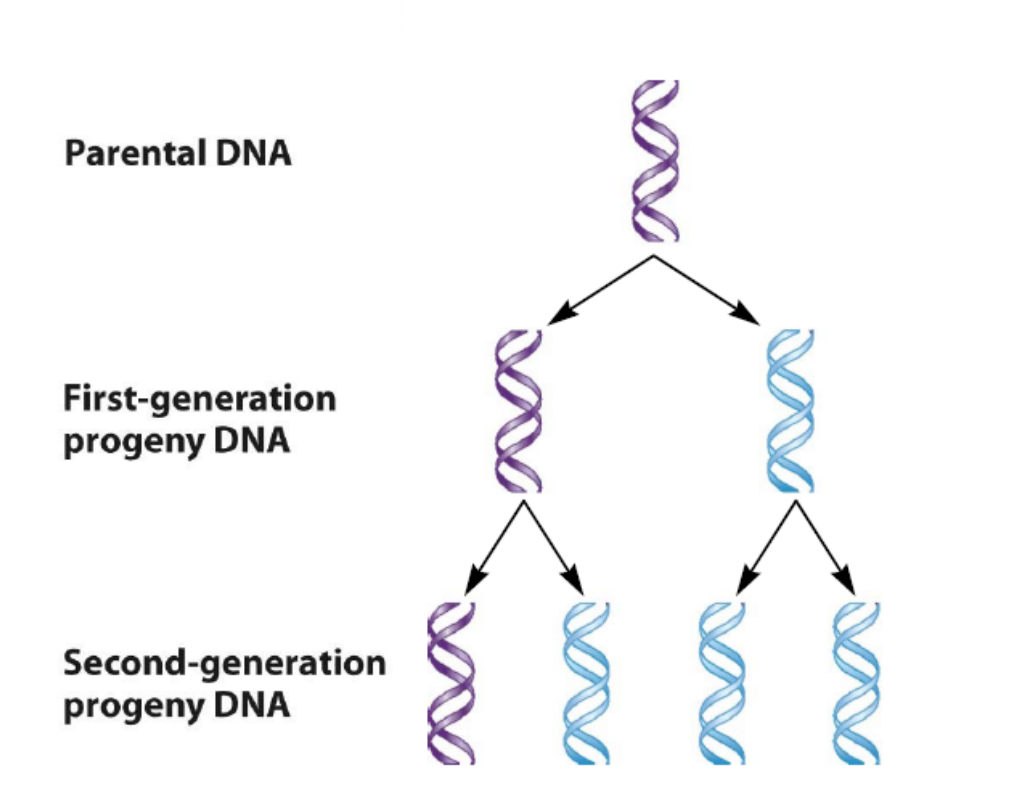
2. Dispersive model 分散模型
Parental and daughter DNA are interspersed in both strands following replication
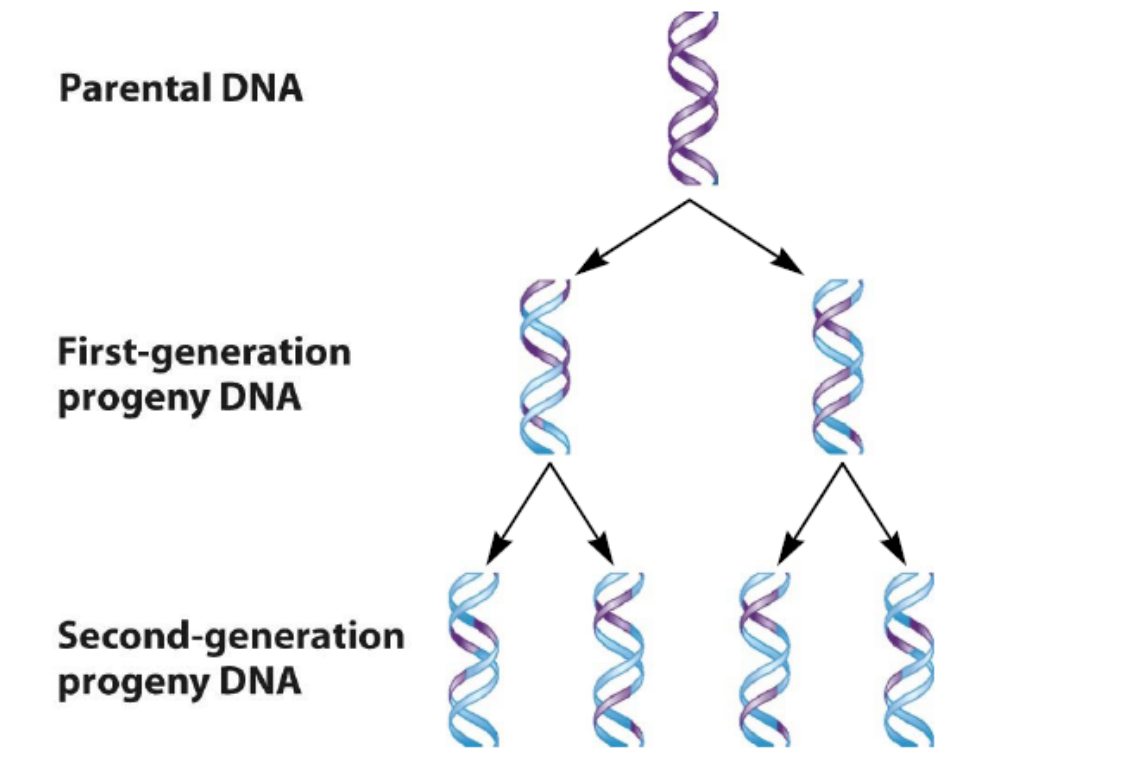
3. Semiconservative model 半保留复制模型
The double-stranded DNA contains one parental and one daughter strand following replication
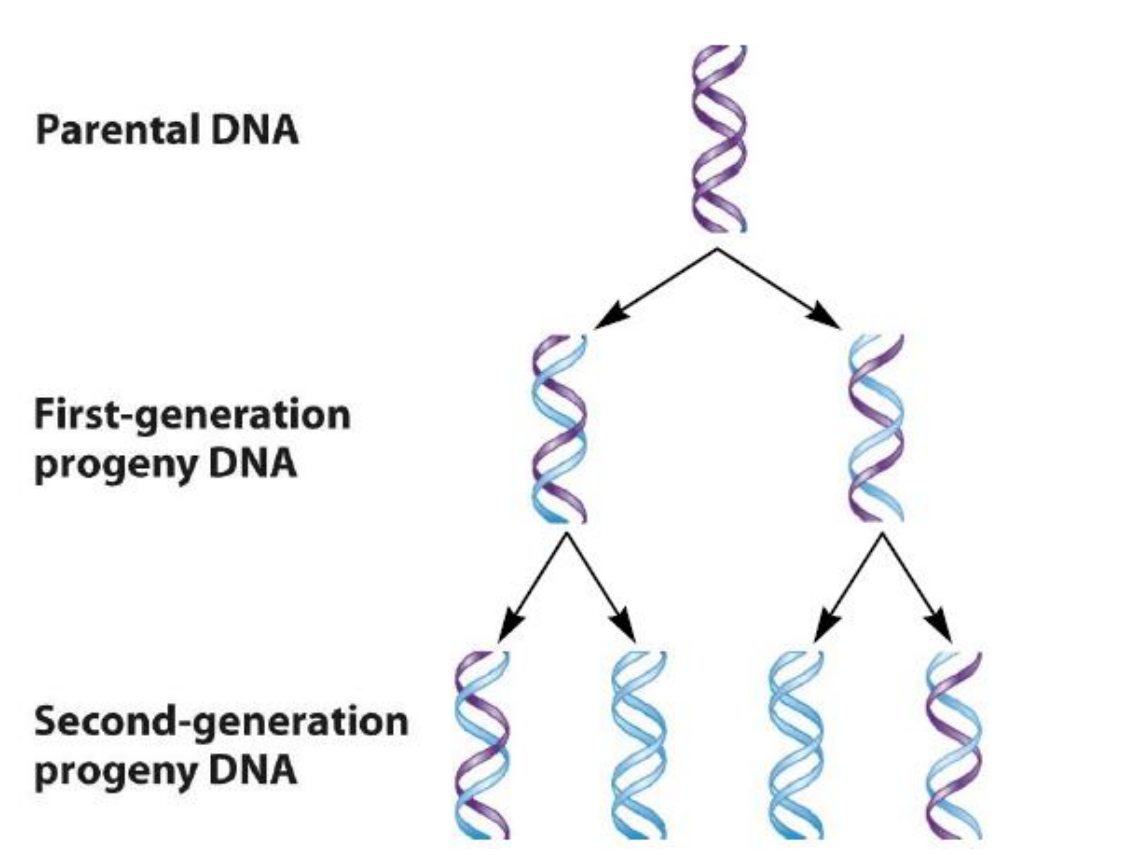
Meselson–Stahl experiment
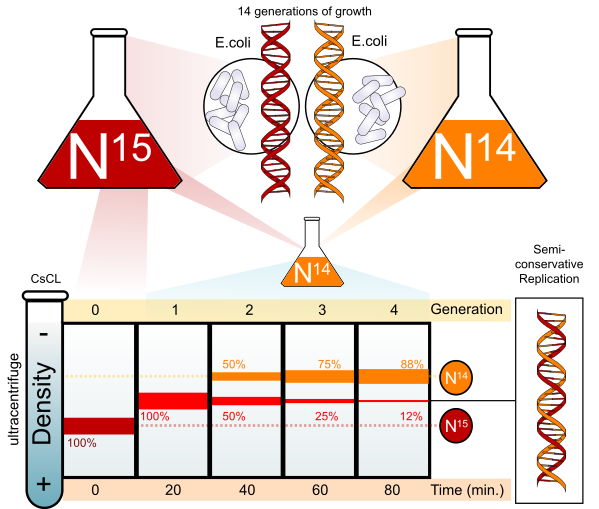
The Characteristic of DNA Replication
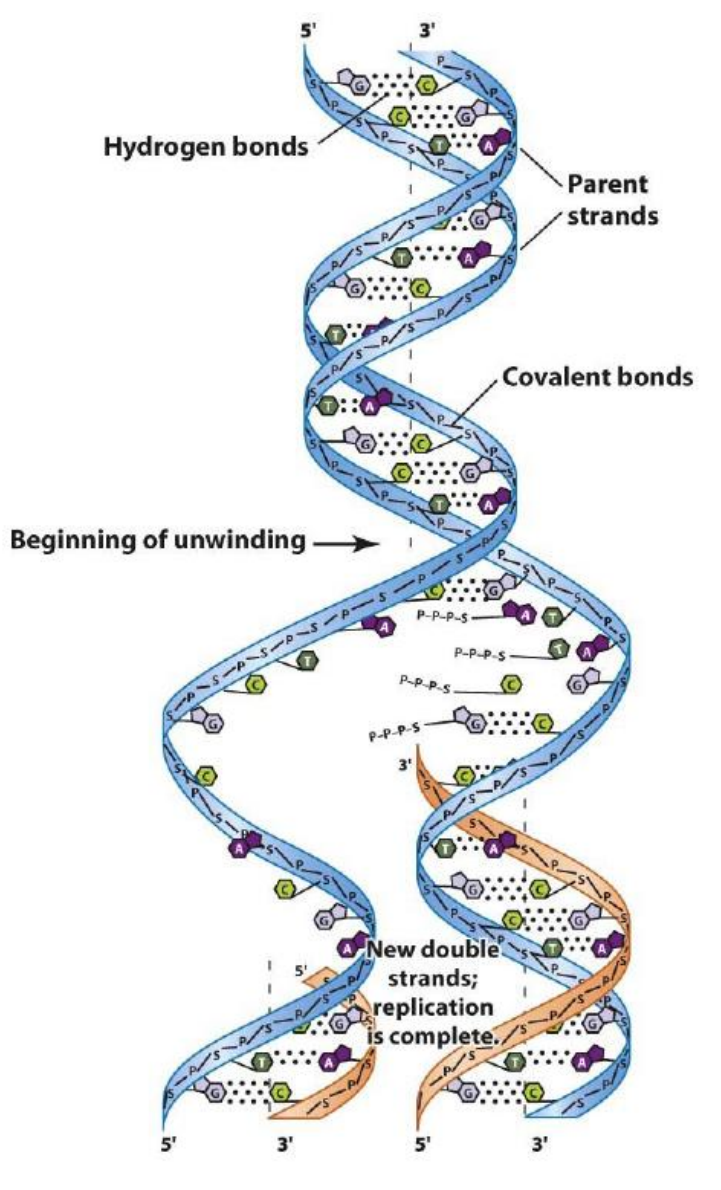
DNA replication relies on the complementarity of DNA strands
- The AT/GC rule (Chargaff’s rule)
The process can be summarized as such:
- The two DNA strands come apart
- Each serves as a template strand for the synthesis of new strands
- The two newly-made strands = daughter strands
- The two original ones = parental strands
二、DNA Replication in Molecular Level
1. DNA replication requires
Coordination with cell division
A special DNA sequence termed the Origin of Replication
- Origin of Replication is site of synthesis initiation and point of control for whole process
DNA polymerases - enzymes to synthesize actual DNA molecule
Accessory enzymes (gyrases, helicases, ligases etc.)
2. DNA replication occurs in three stages
- Initiation
- Elongation
- Termination
3. Different Strand
| Lagging Strand | Discontinuous Replicate |
|---|---|
| Leading Strand | Continuous Replicate |
DNA replication: Initiation
1. Origins of Replication
E.coli
Only one origin of replication in E. coli (but many in eukaryotes)
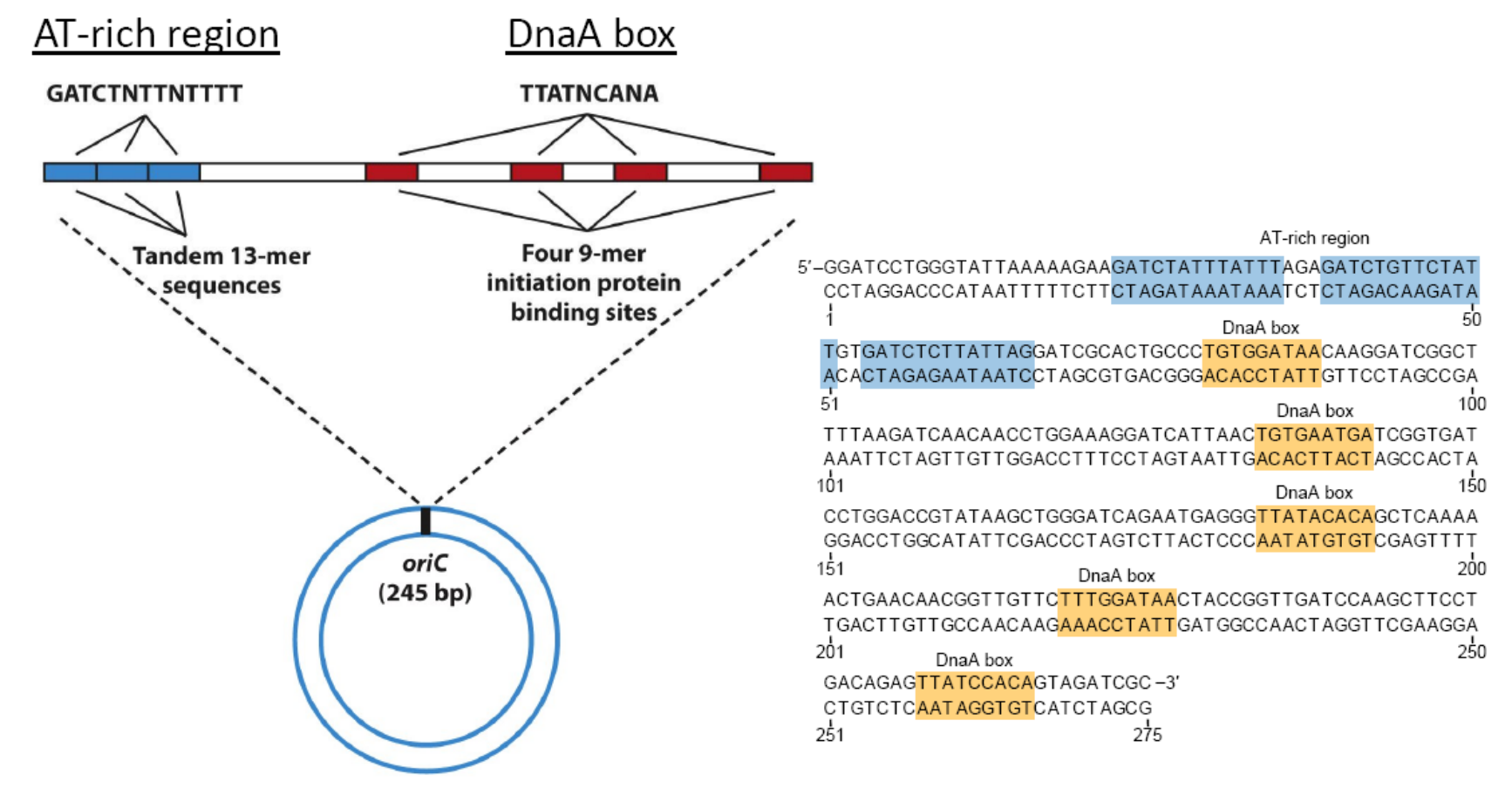
The origin of replication in E. coli is termed oriC
- origin of Chromosomal replication
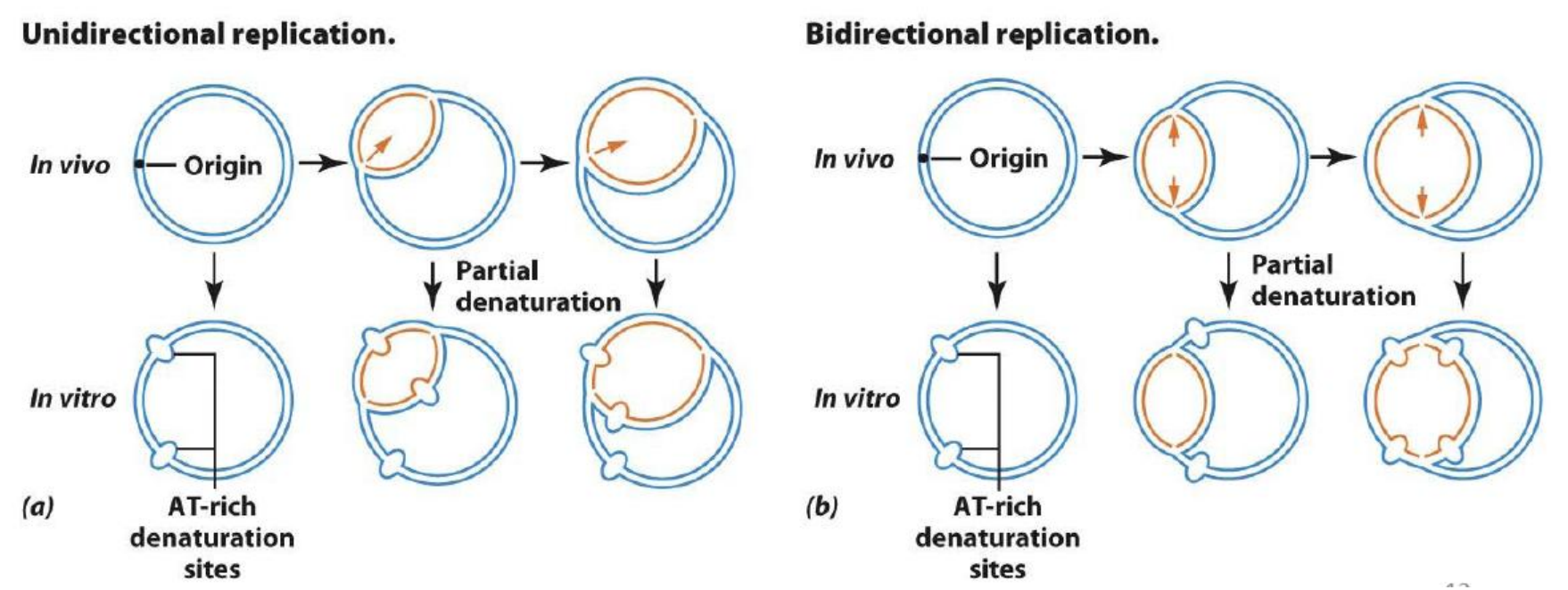
E.coli的复制是双向的(Bidirectional)
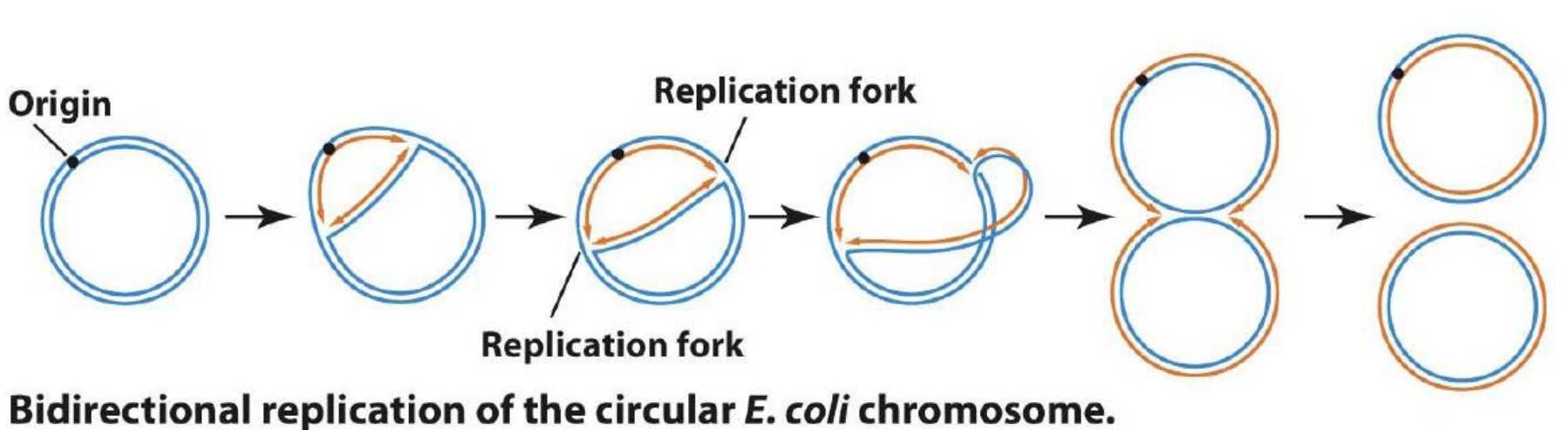
The characteristic of this replication is called θ replication
- 因为复制的时候形成了类似θ的结构
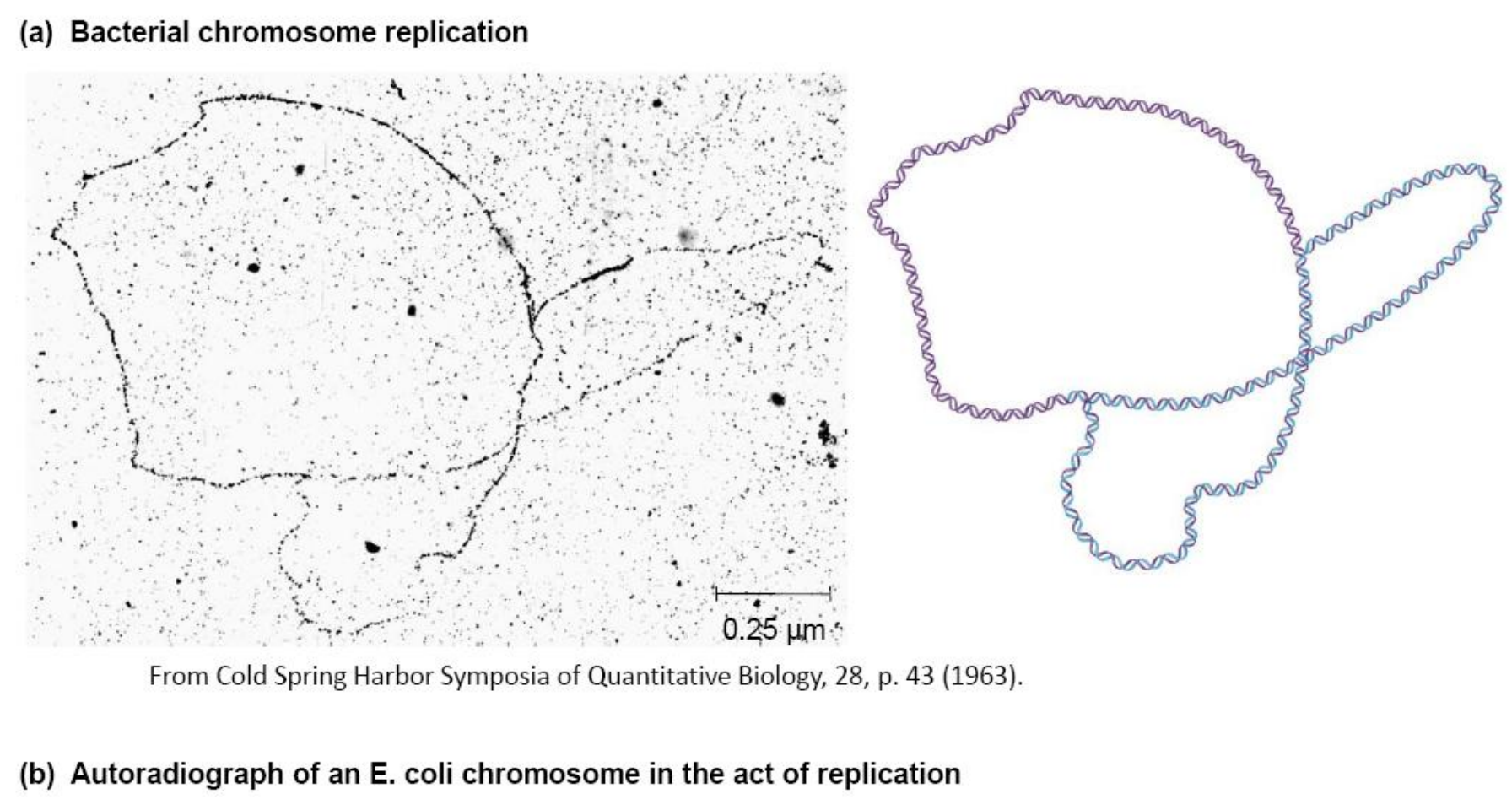
2. Proof of Bidirectional Replication
Expose to pH 11.05 for 10 minutes AT-rich clusters denature to form denaturation bubbles
3. DNA Unwinding 解旋
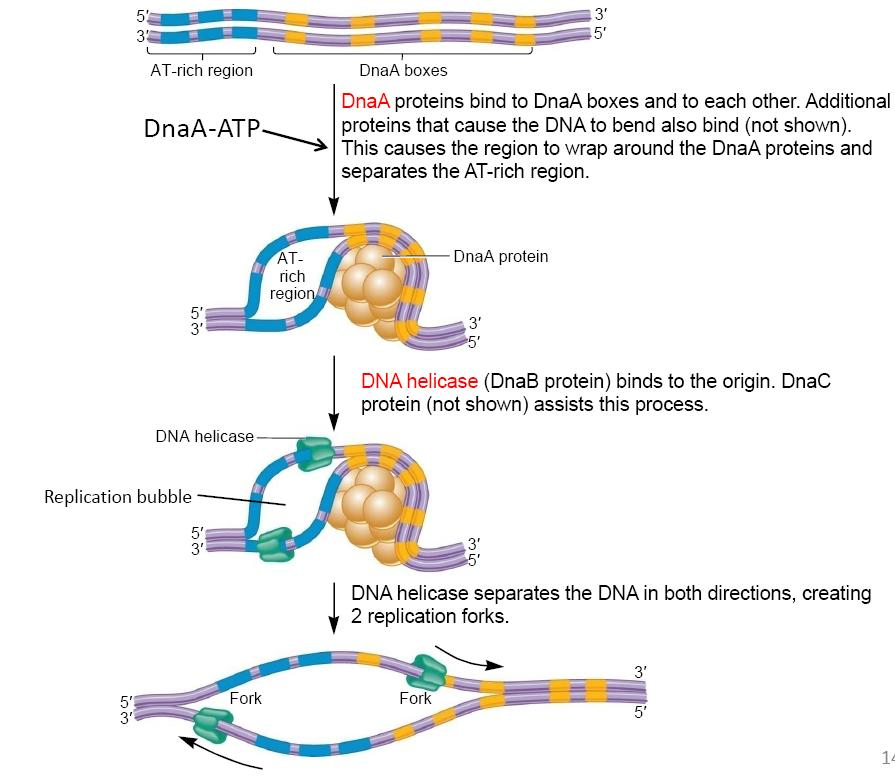
DnaA proteins bind to DnaA boxes and to each other. Additional proteins that cause the DNA to bend also bind (not shown).
This causes the region to wrap(缠绕) around the DnaA proteins and separates theAT-rich region.
DNA helicase(DNA 解旋酶) (DnaB protein) binds to the origin. DnaC protein (not shown) assists this process.
DNA helicase separates the DNA in both directions, creating 2 replication forks.
The DNA helicase travels from 5’ to 3’ position in bacteria DNA, this is bidirectional replication.
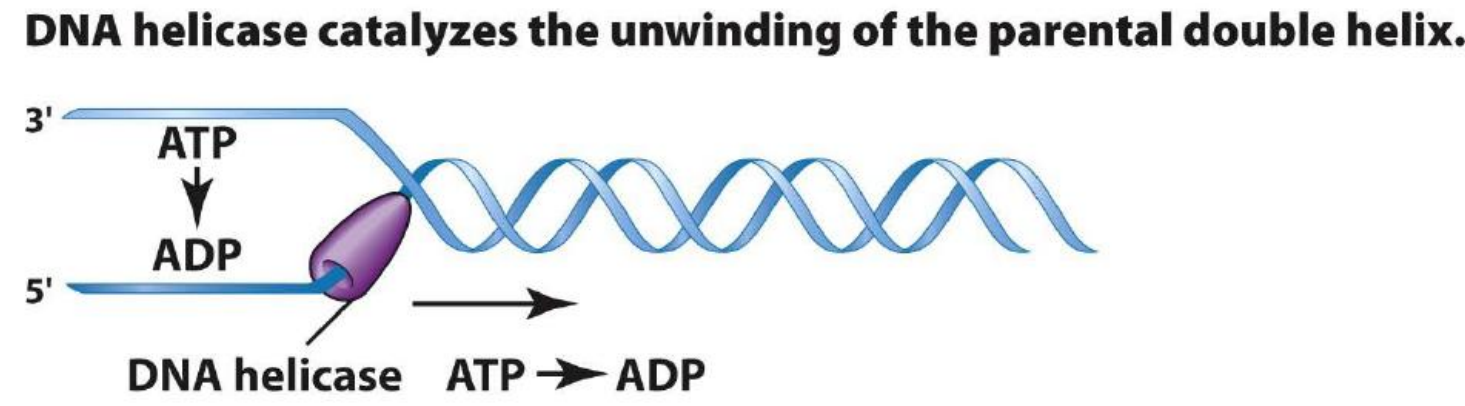
DNA Helicase
They are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two annealed nucleic acid strands (i.e., DNA, RNA, or RNA-DNA hybrid) using energy derived from ATP hvdrolysis.
(2) Structure And Moving Direction in Different Species
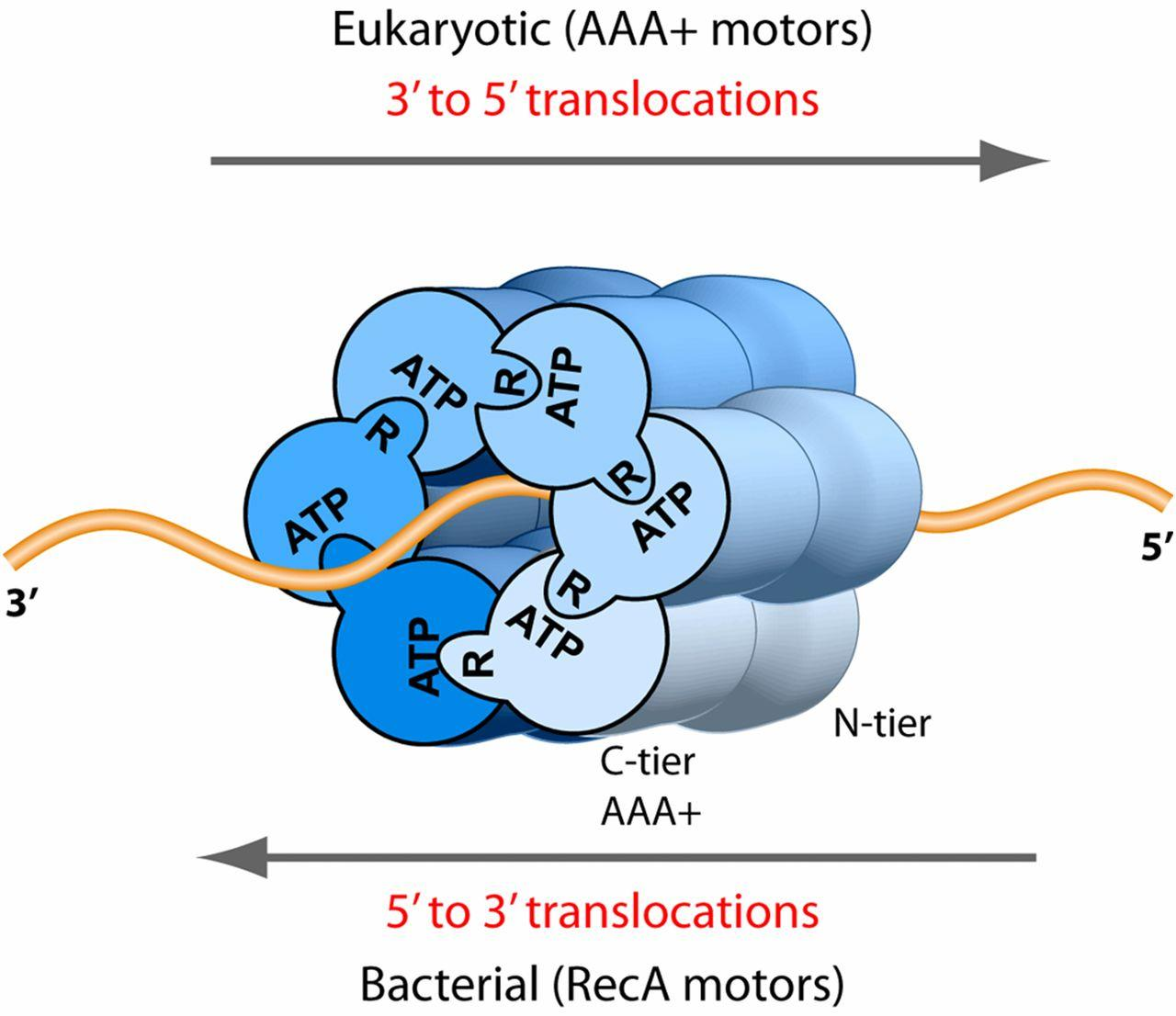
| Eukaryote | 3’ to 5’ | 6 different subunit |
|---|---|---|
| Archaea | 3’ to 5’ | 6 same subunit |
| Bacteria | 5’ to 3’ | 6 same subunit |
Single-Strand DNA Binding (SSB) Protein
Single-strand DNA-binding (SSB) protein keeps the unwound strands in an extended form for replication
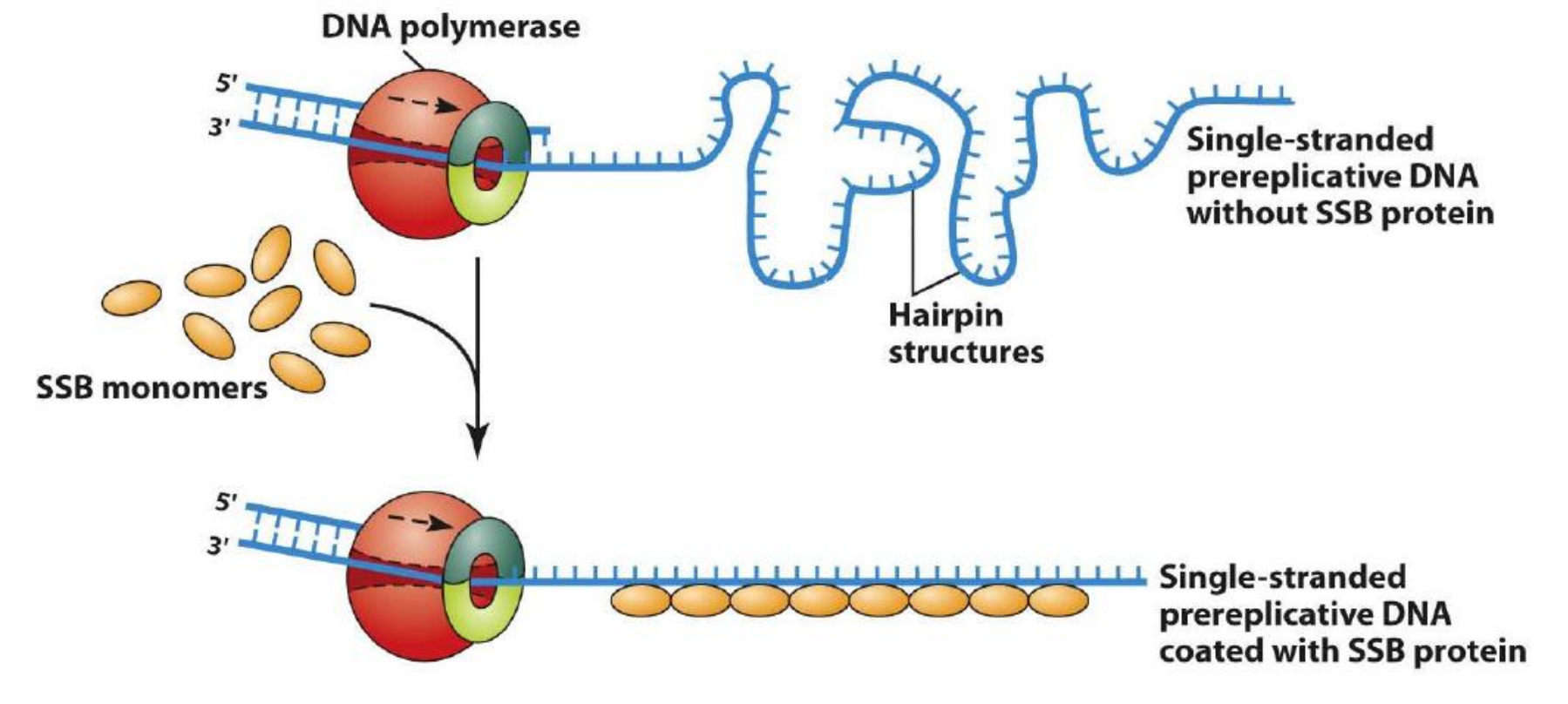
(1) SSB protein
SSB are essential proteins found in all domains of life.
First, ssDNA is thermodynamically less stable than dsDNA, which leads to spontaneous formation of duplex secondary structures that impede genome maintenance processes.
Second, relative to dsDNA, ssDNA is hypersensitive to chemical and nucleolytic attacks that can cause damage to the genome.
Supercoiling
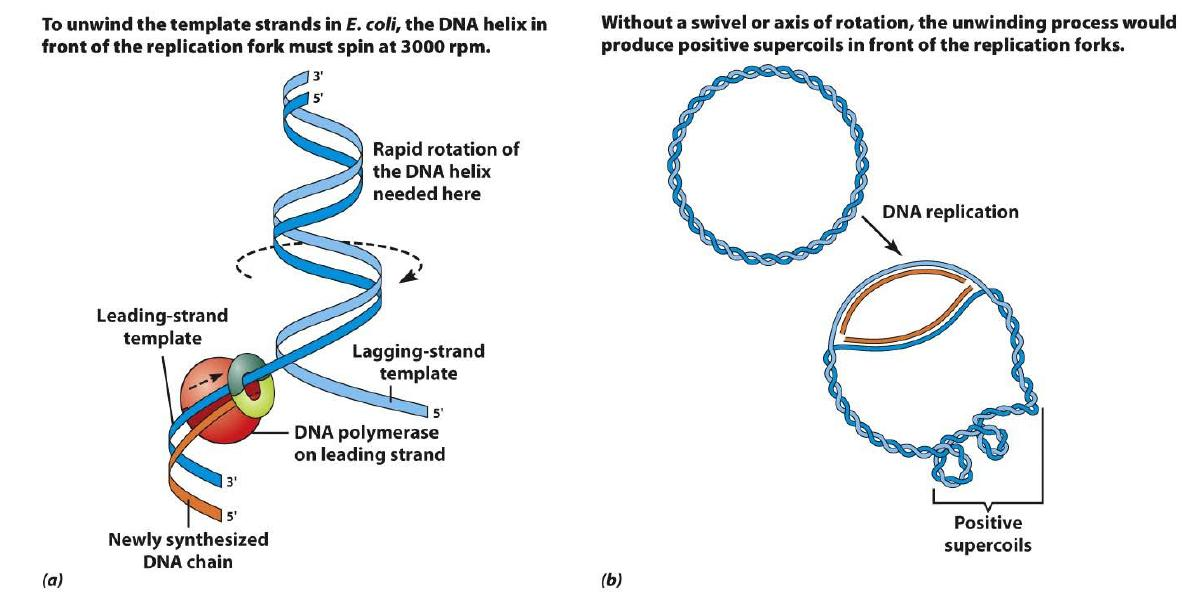
(a) To unwind the template strands in E. coli, the DNA helix in front of the replication fork must spin at 3000 rpm.
(b) Without a swivel or axis of rotation, the unwinding process would produce positive supercoils in front of tlra replication fork.
DNA topoisomerases (Gyrase)
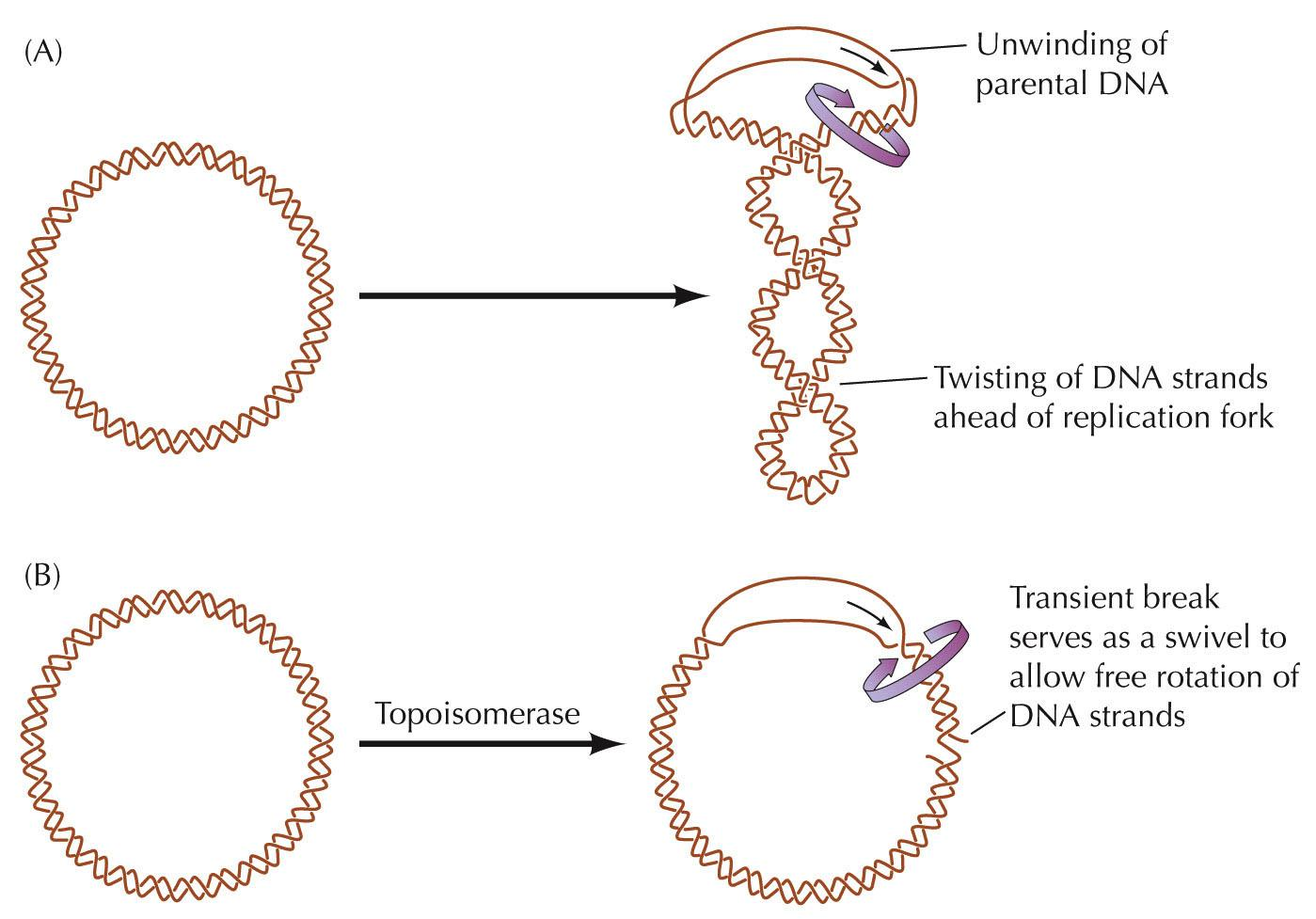
DNA replication: Elongation
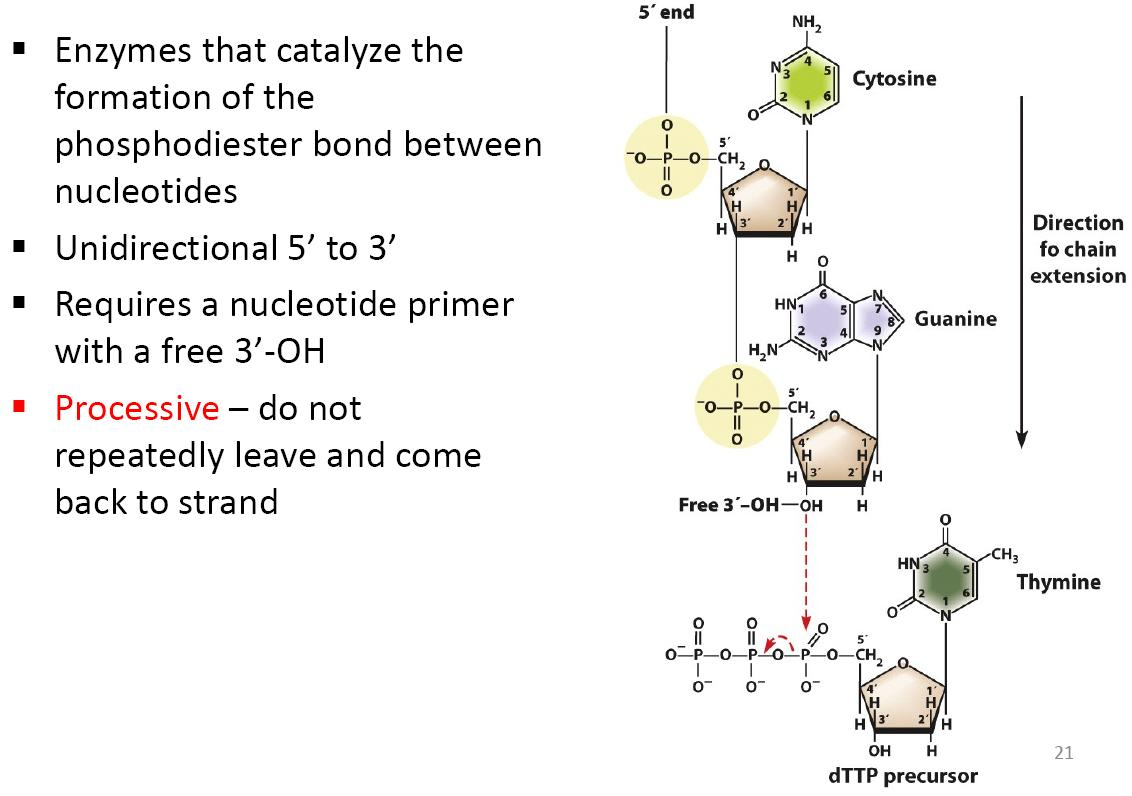
Enzymes that catalyze the formation of the phosphodiester bond between nucleotides
Unidirectional 5’ to 3’
Requires a nucleotide primer with a free 3’-OH
Processive 一 do not repeatedly leave and come back to strand
1. Discontinuous and Continuous Synthesis
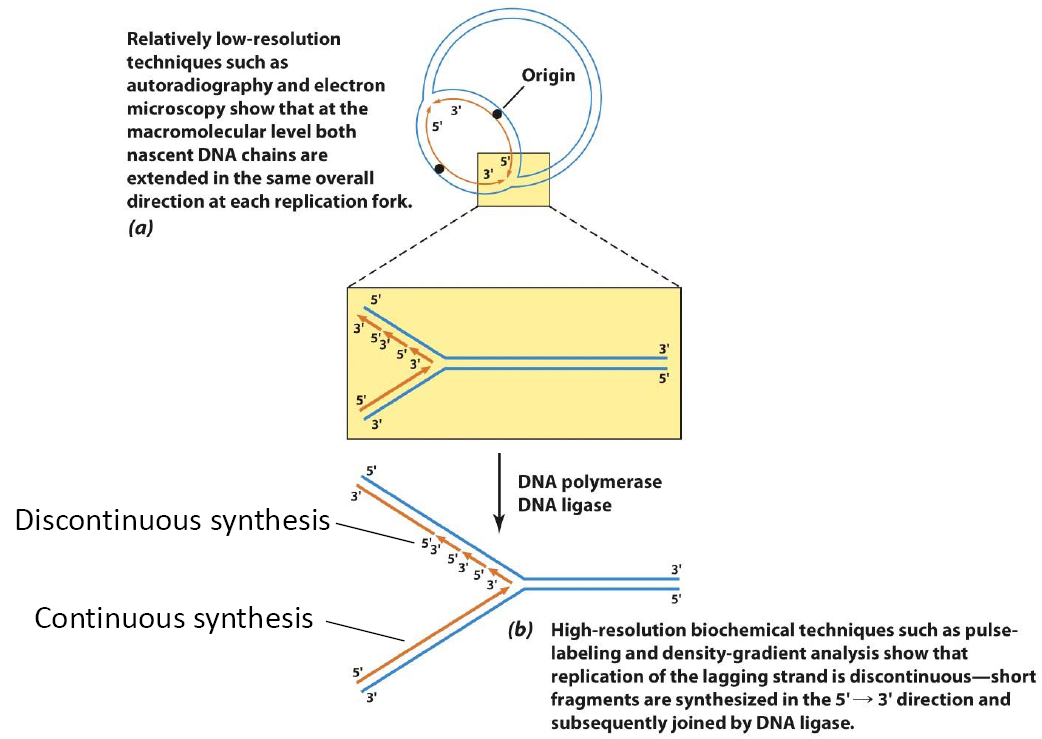
Relatively low-resolution techniques such as autoradiography and electron microscopy show that at the macromolecular level both nascent DNA chains are extended in the same overall direction at each replication fork.
High-resolution biochemical techniques such as pulse- labeling and density-gradient analysis show that replication of the lagging strand is discontinuous—short fragments are synthesized in the 5’— 3’ direction and subsequently joined by DNA ligase.

DNA pol: Need primer
RNA pol: Doesn’t need primer
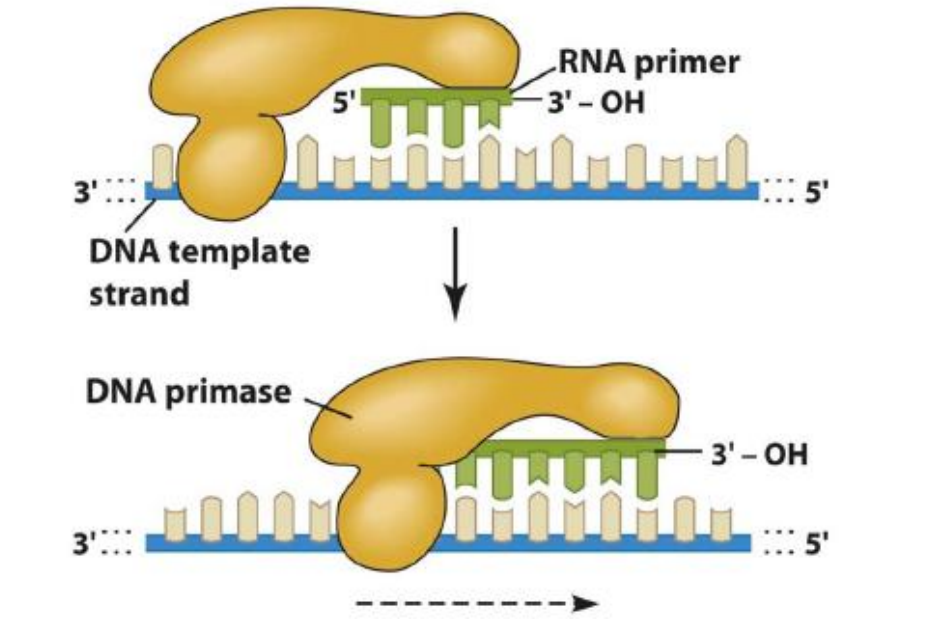
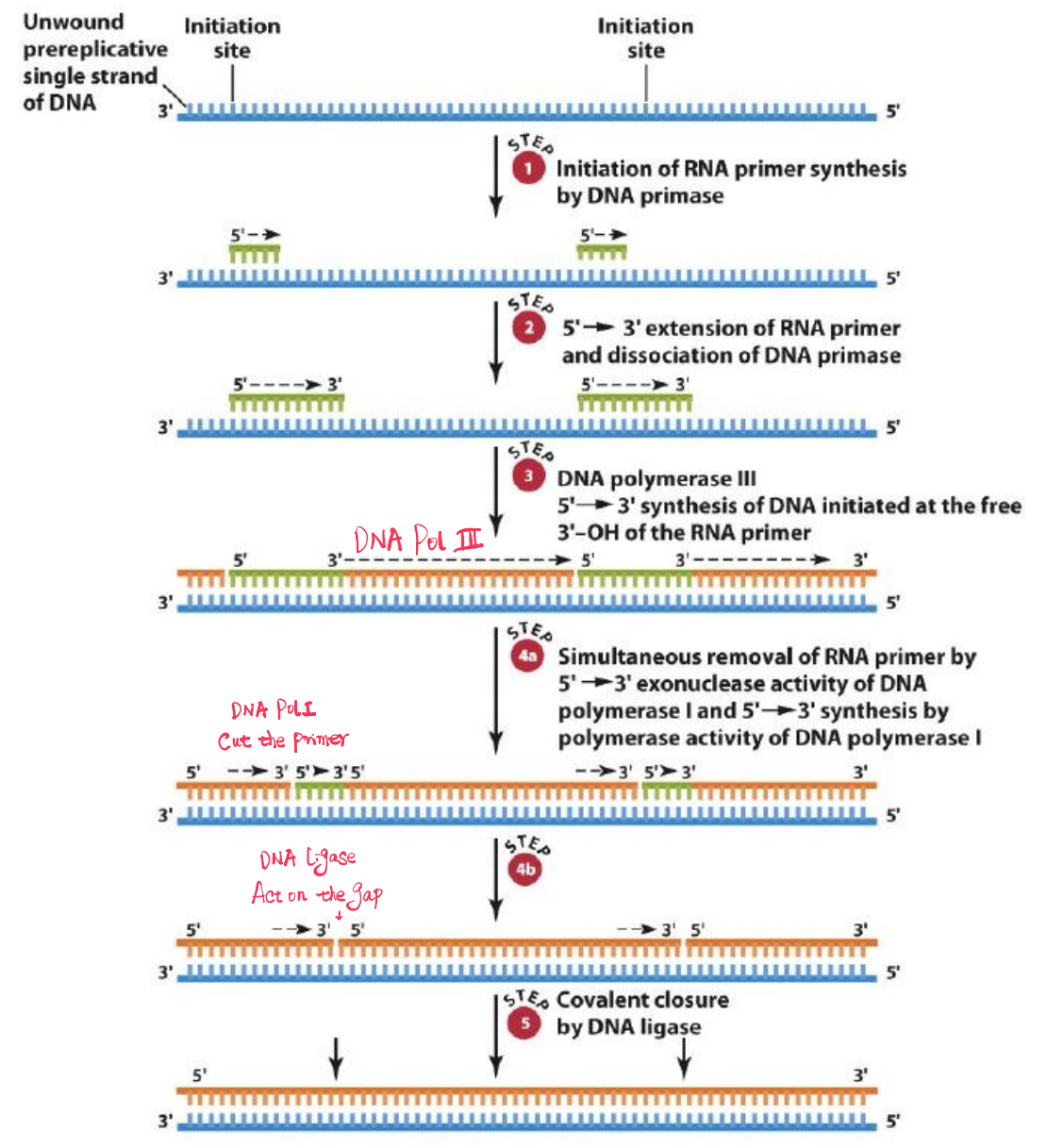
2. Initiation of DNA Chains with RNA Primers
Primase creates a short RNA primer to which the DNA polymerase can then add more deoxynucleotide triphosphates (dNTPs)
DNA primase creates primers for both strands
- One primer for strand being synthesized into the fork - leading strand
- Multiple primers for strand being synthesized away from the fork - Lagging strand
- Many short RNA-DNA molecules on lagging strand known as Okazaki fragments
3. DNA Polymerases
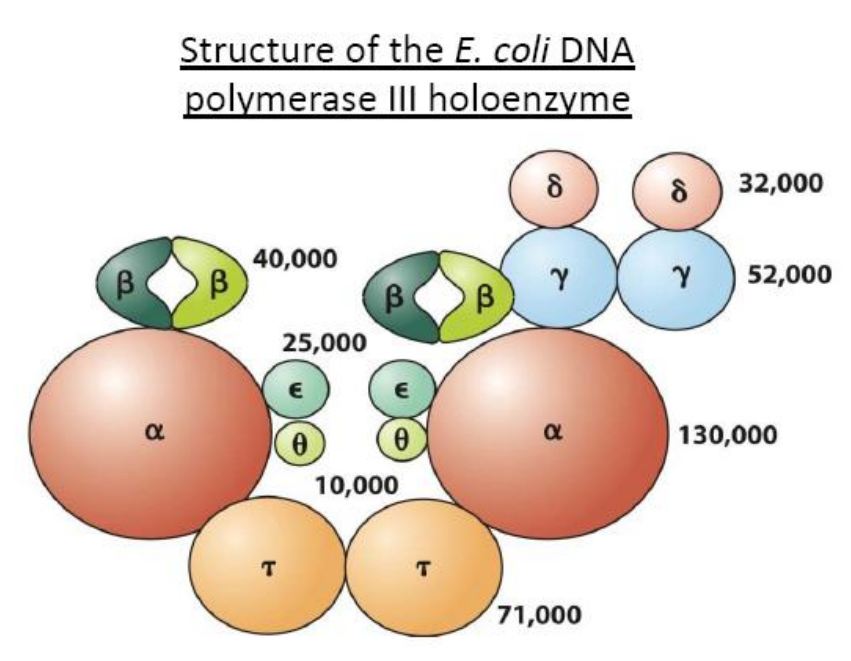
DNA Polymerases Ⅲ
DNA Polymerase III synthesizes the DNA using these primers on both leading and lagging strands.
But can not remove RNA primer from Okazaki fragments
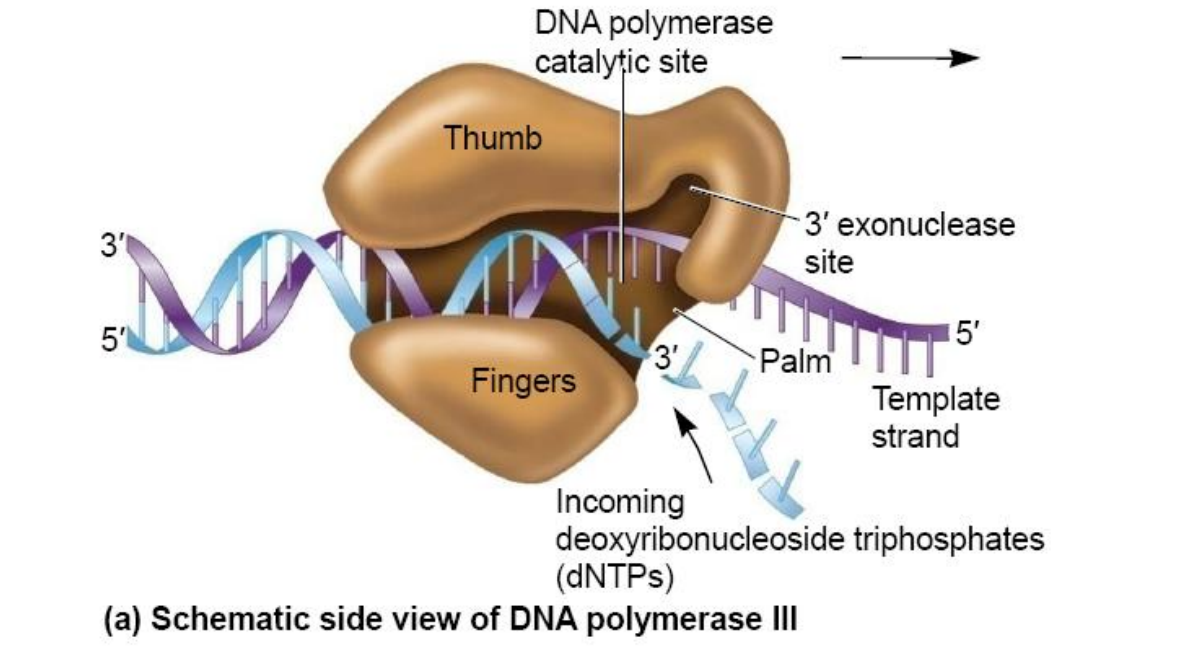
Only the DNA pol III is the replicase
The others participate in repair of damaged DNA.
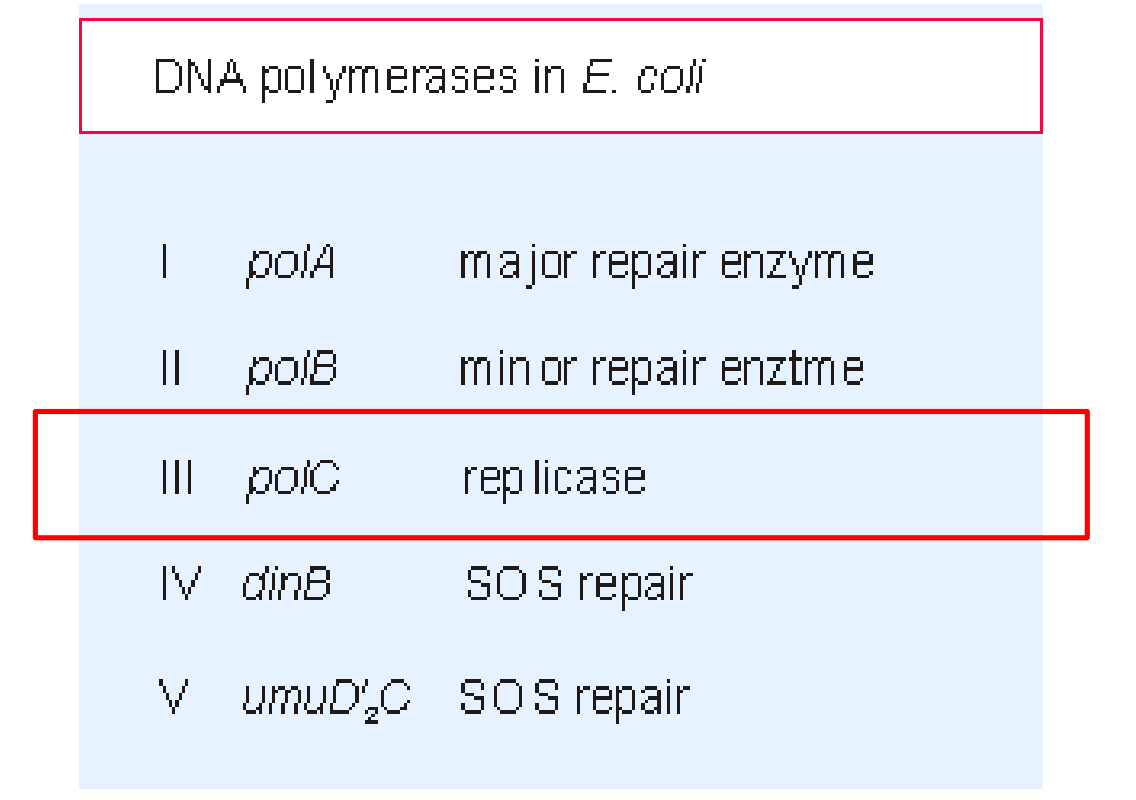
(1) Structure of DNA pol III in E.coli
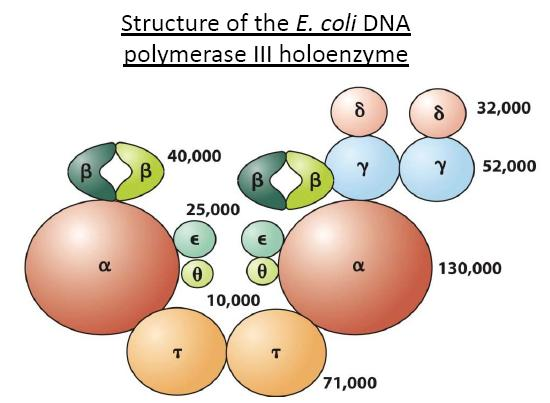
DNA Polymerases Ⅰ
DNA polymerase I
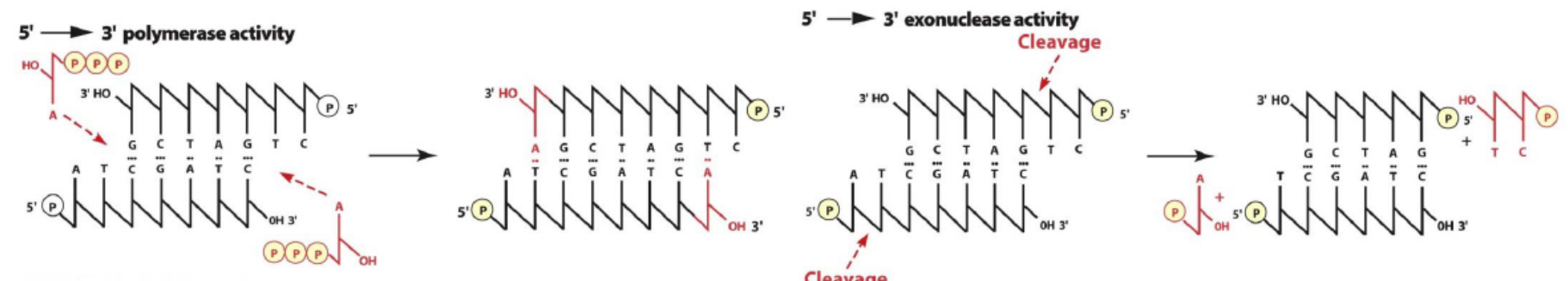
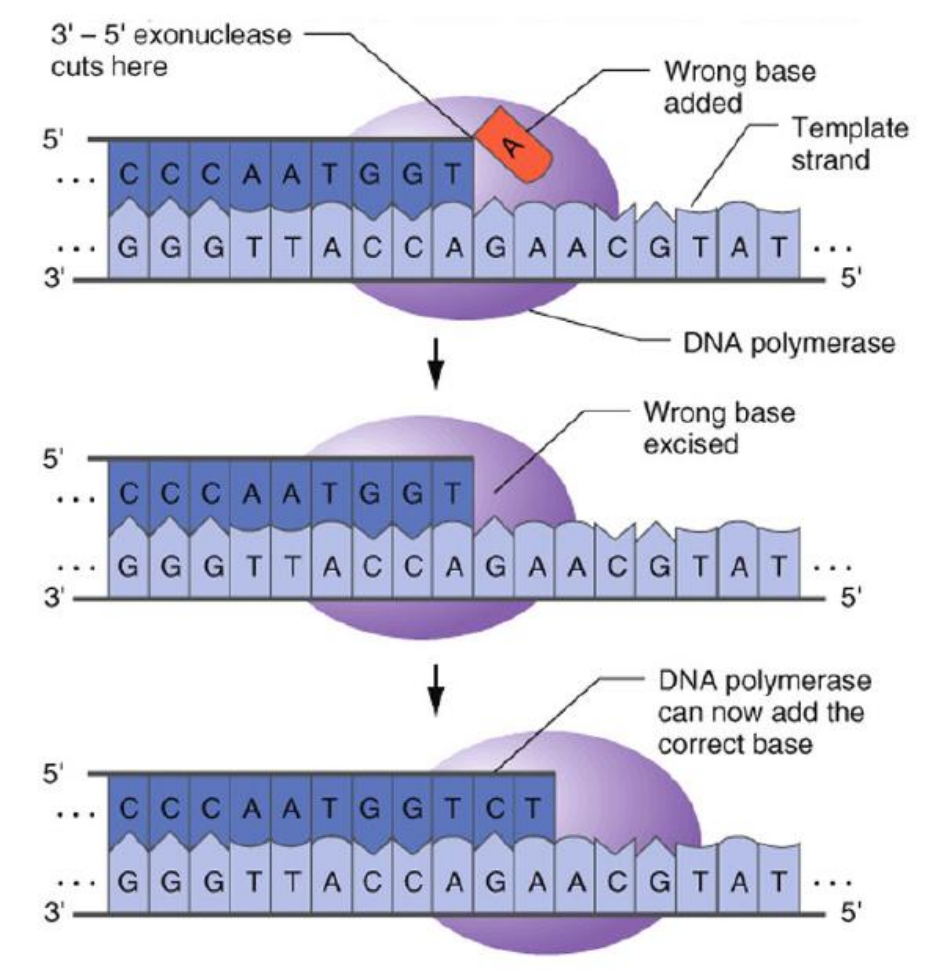
- Also has 5’ to 3’ exonuclease(核酸外切酶) activity
- Primarily acts on only the lagging strand to remove RNA from Okazaki fragments
- Uses 5’-3‘ polymerase activity to replace RNA with DNA
Eukaryotes Contain Several Different DNA Polymerases
Mammalian cells contain well over a dozen different DNA polymerases
Four: alpha (α), delta (δ), epsilon (ε) and gamma (γ) have the primary function of replicating DNA
α, δ and ε → Nuclear DNA
γ → Mitochondrial DNA

DNA pol α is the only polymerase to associate with primase
- The DNA pol α/primase complex synthesizes a short RNA-DNA hybrid
- This is used by DNA pol δ or ε for the processive elongation of the leading and lagging strands
The exchange of DNA pol α, δ and ε is called a polymerase switch
- It occurs only after the RNA-DNA hybrid is made
4. DNA Ligase
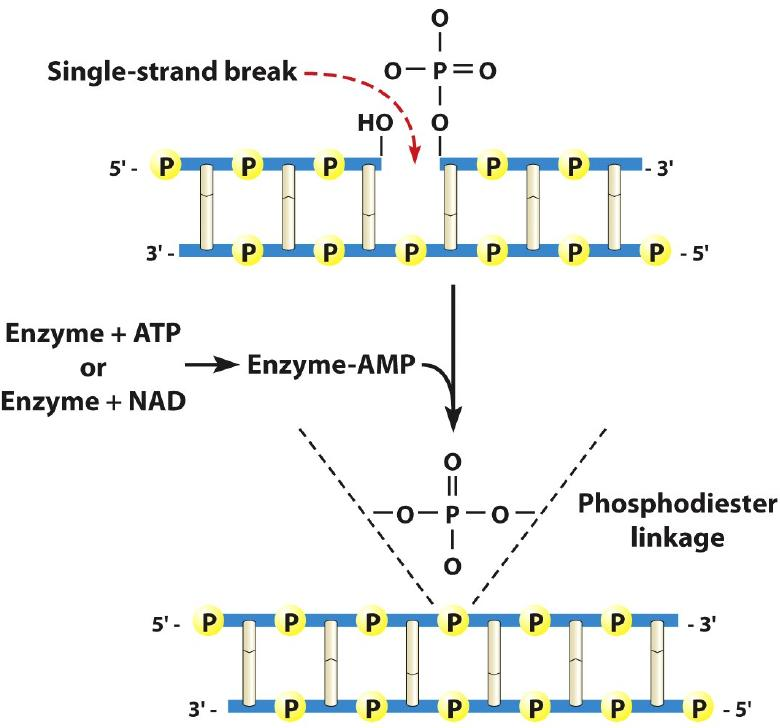
5. Summary of Elongation
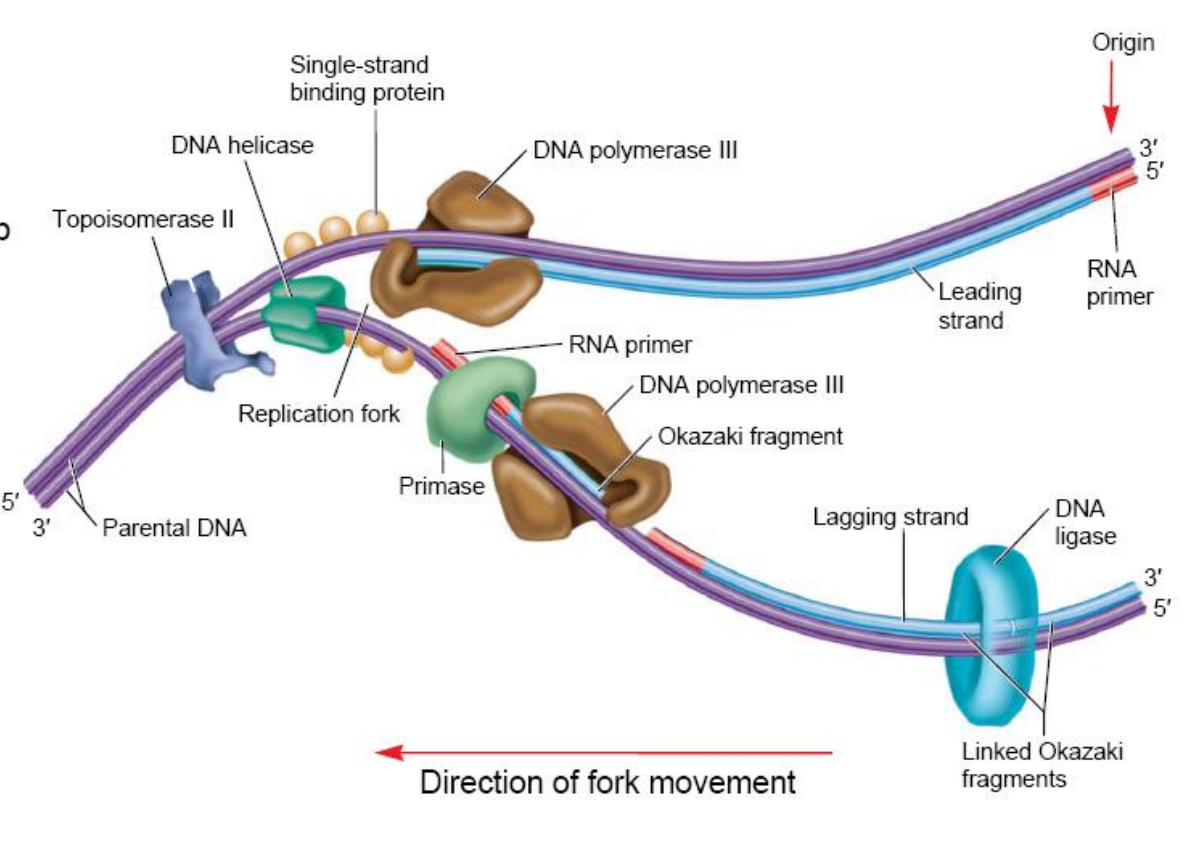
- DNA helicase breaks the hydrogen bonds between the DNA strands.
- Topoisomerase alleviates positive supercoiling.
- Single-strand binding proteins keep the parental strands apart.
- Primase synthesizes an RNA primer.
- DNA polymerase III synthesizes a daughter strand of DNA.
- DNA polymerase I excises the RNA primers and fills in with DNA (not shown).
- DNA ligase covalently links the Okazaki fragments together
DNA Replication Complexes
DNA helicase and primase are physically bound to each other to form a complex called the primosome(引发体)
引发体(Primosome)是在DNA复制过程当中,引发合成各个冈崎片段时所需要的多蛋白复合物(DNA复制过程中合成RNA引物的多种蛋白质复合体)。主要是由引物酶和DNA解旋酶等成份组成。引发体沿着不连续合成的DNA的那一条链(复制叉)移动,移动过程需要三磷酸腺苷(ATP)供能,移动方向与RNA及DNA的合成方向相反。
- This complex leads the way at the replication fork
The primosome is physically associated with the DNA polymerase holoenzyme forming the replisome(复制体)
复制体(英语:Replisome)是参与DNA复制的蛋白质复合物(执行DNA复制功能的多种蛋白质复合体),其中至少含有DNA聚合酶及引发体(primosome,引发酶与其他分子的复合物)。以大肠杆菌为例,此类细菌的复合体包含两组DNA聚合酶III全酶(DNA polymerase III holoenzyme)、引发酶与螺旋酶。
复制体作用时位于复制叉附近,沿着复制叉开启的方向移动,合成DNA。
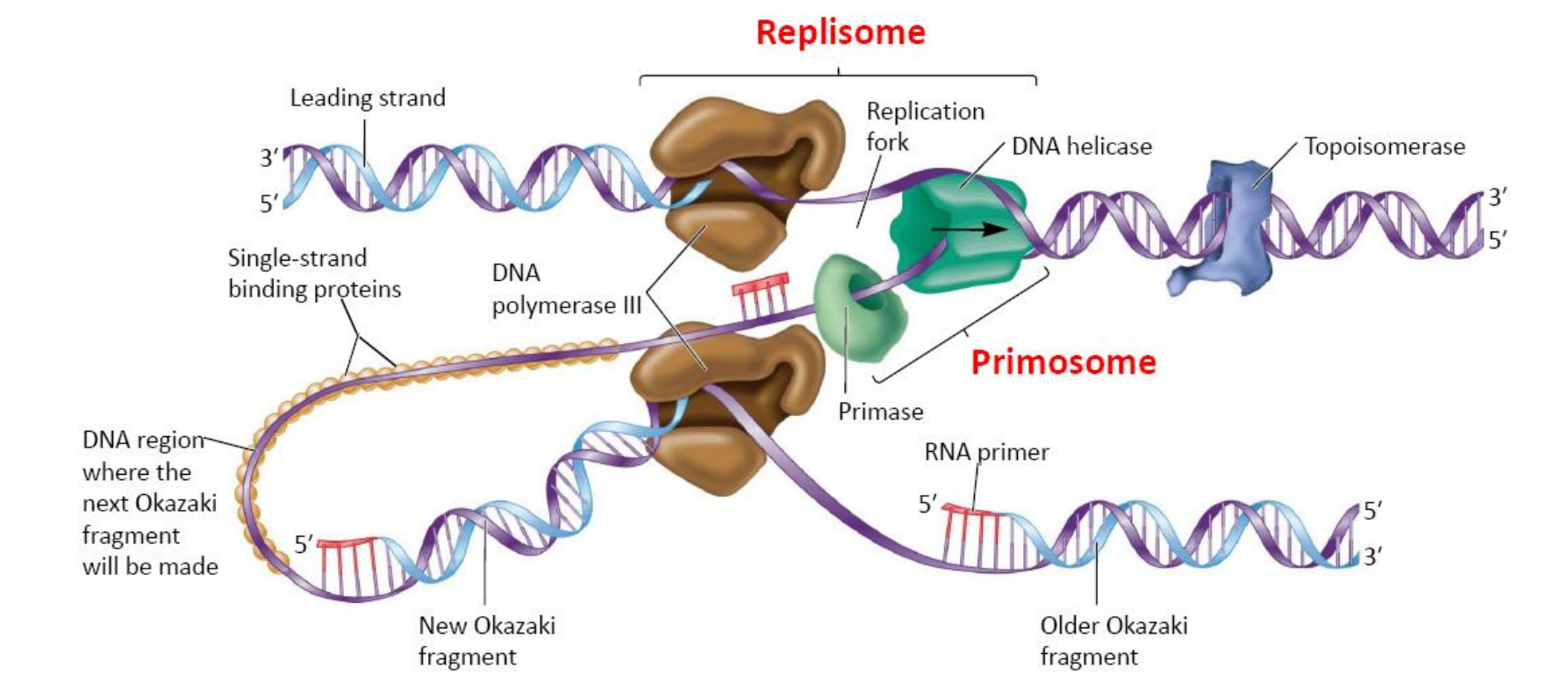
Two DNA pol III proteins act in concert to replicate both the leading and lagging strands
- The two proteins form a dimeric DNA polymerase that moves as a unit toward the replication fork
DNA replication: Termination
In E. coli, opposite to oriC is a pair of termination sequences called ter sequences. These are designated T1 and T2,
- T1 stops counterclockwise forks, T2 stops clockwise
The protein tus (**t**ermination **u**tilization **s**ubstance) binds to these sequences
- It can then stop the movement of the replication forks
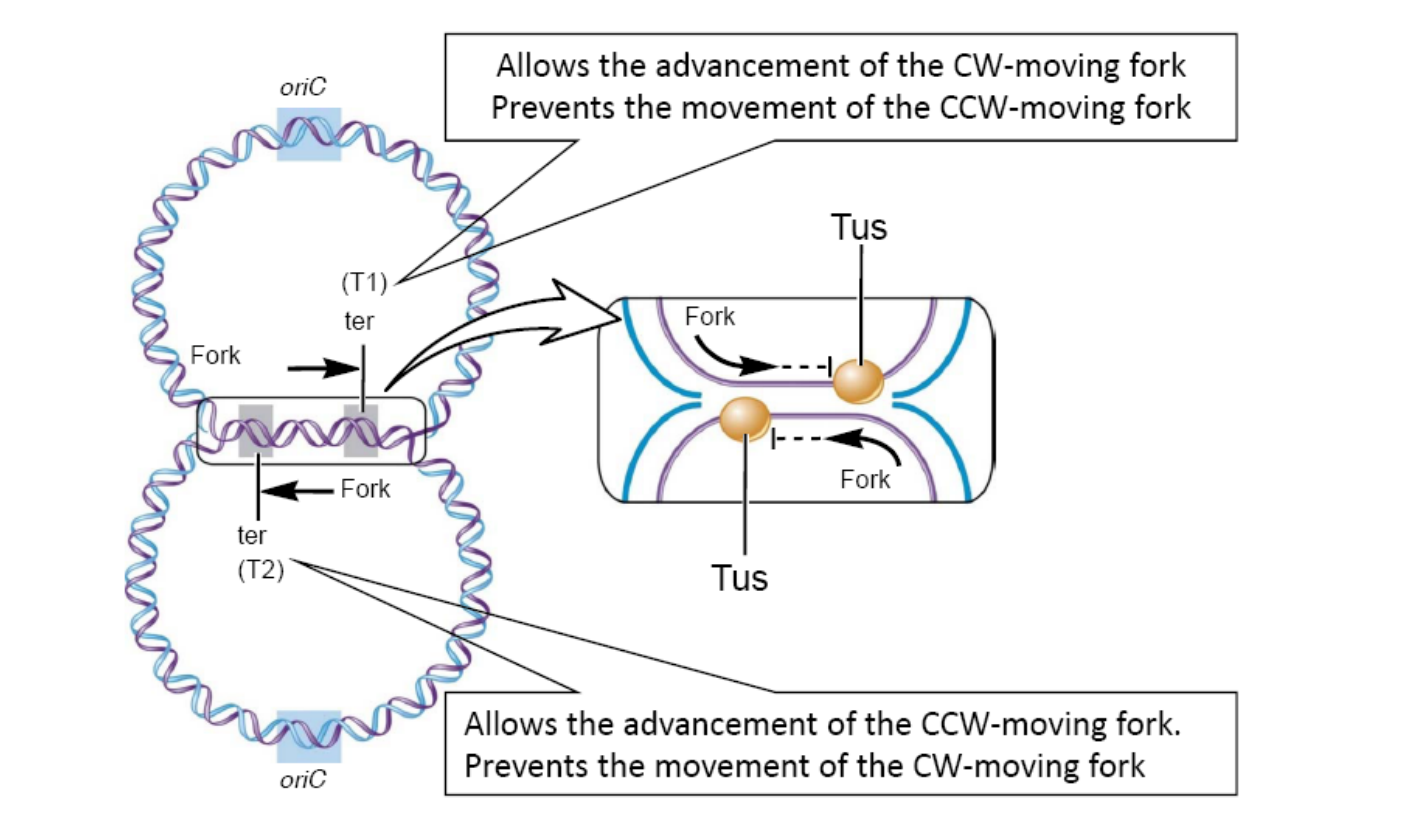
DNA replication ends when oppositely advancing forks meet (usually at T1 or 72)
DNA replication often results in two intertwined(纠缠;编结) molecules
- Intertwined circular molecules are termed catenanes(索烃)
索烃是一个机械互锁分子,包含有两个或两个以上互锁的大环分子。除非环分子内部的共价键断裂,否则互锁的环不能够分开。索烃的英文名(Catenane)源自拉丁文的Catena,意思是“链”。索烃和其他机械互锁分子有着概念上的联系,例如:轮烷、分子扭结和分子博洛米尼结。
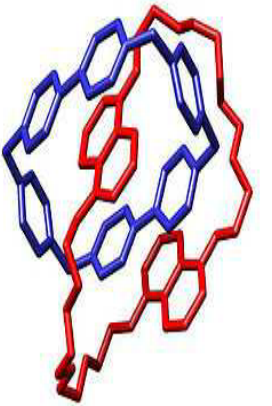
- These are separated by the action of topoisomerases(拓扑异构酶)
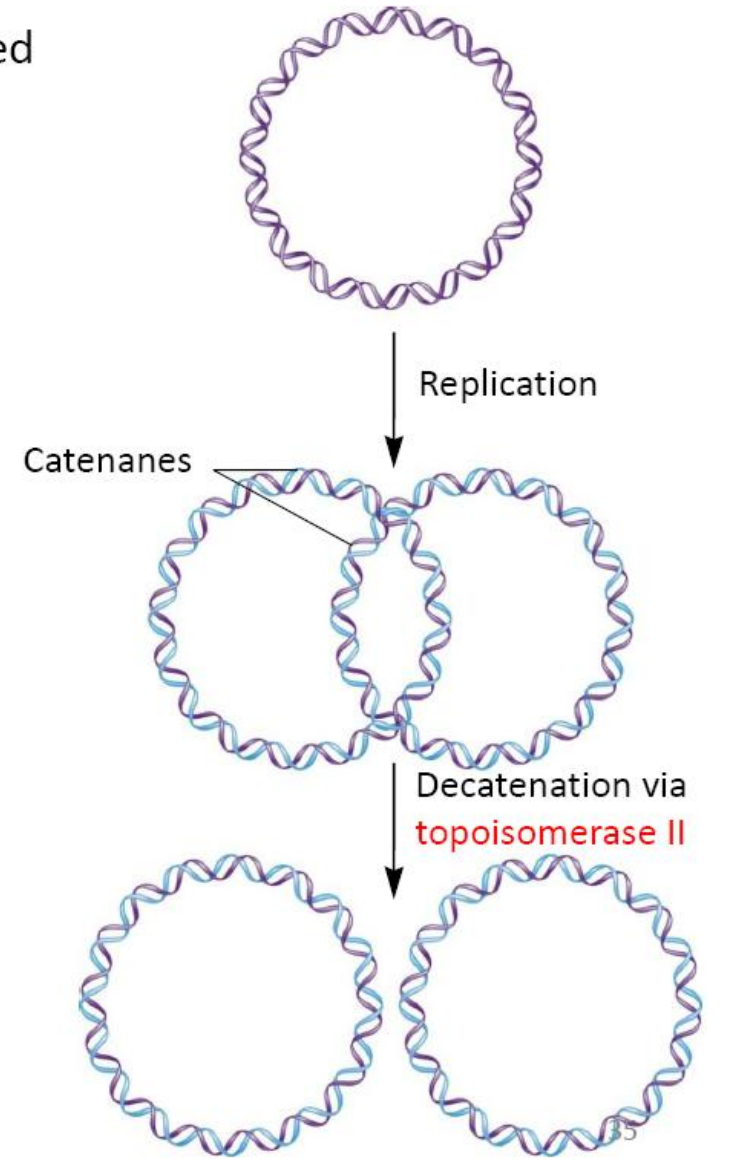
Proofreading 校正
Proofreading activity carry out by the 3’ -5’ exonuclease activities of DNA polymerase
DNA polymerase I of E. coli has build in 3’-5’ exonuclease activities
DNA polymerase III of E coli requires s subunit to carry out 3’-5’exonuclease activities
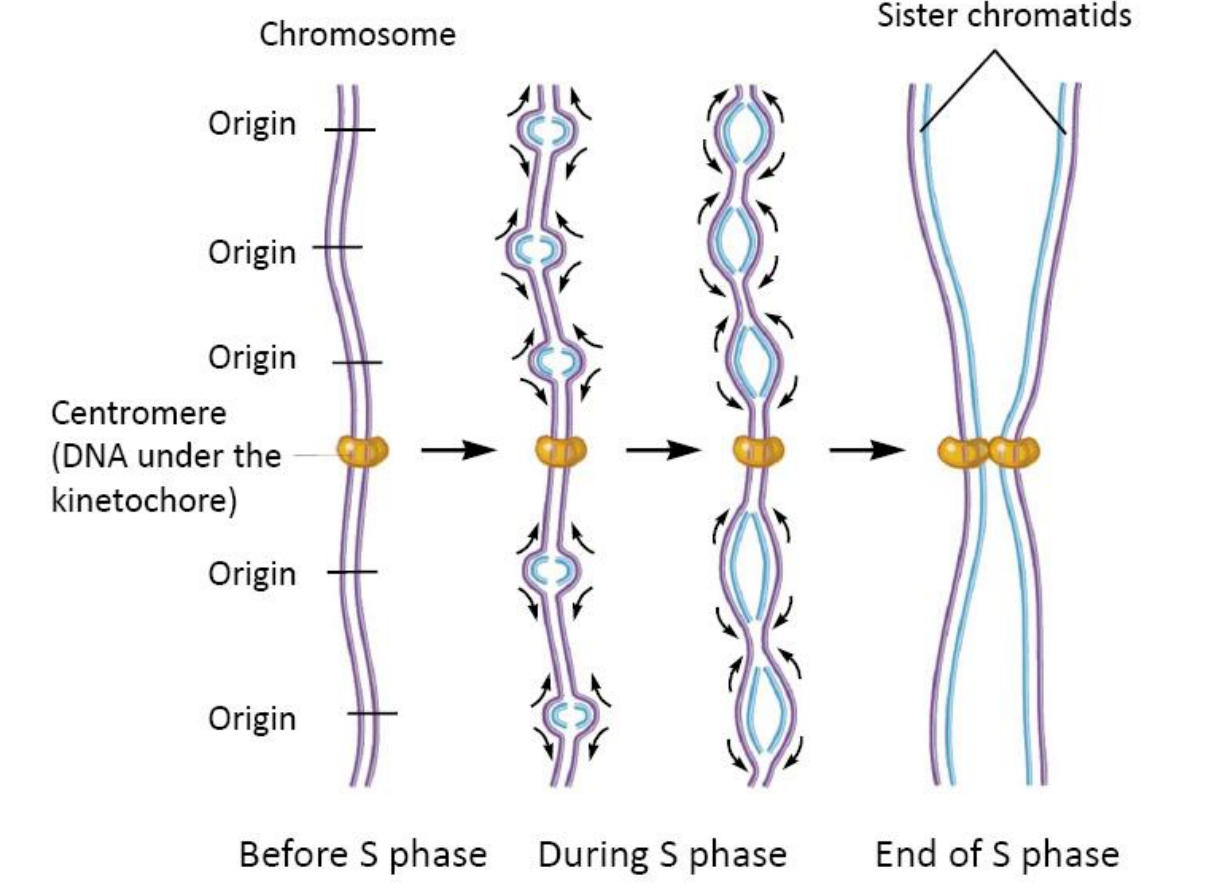
Multiple Origins of Replication
Eukaryotes have long linear chromosomes
- They therefore require multiple origins of replication, To ensure that the DNA can be replicated in a reasonable time.
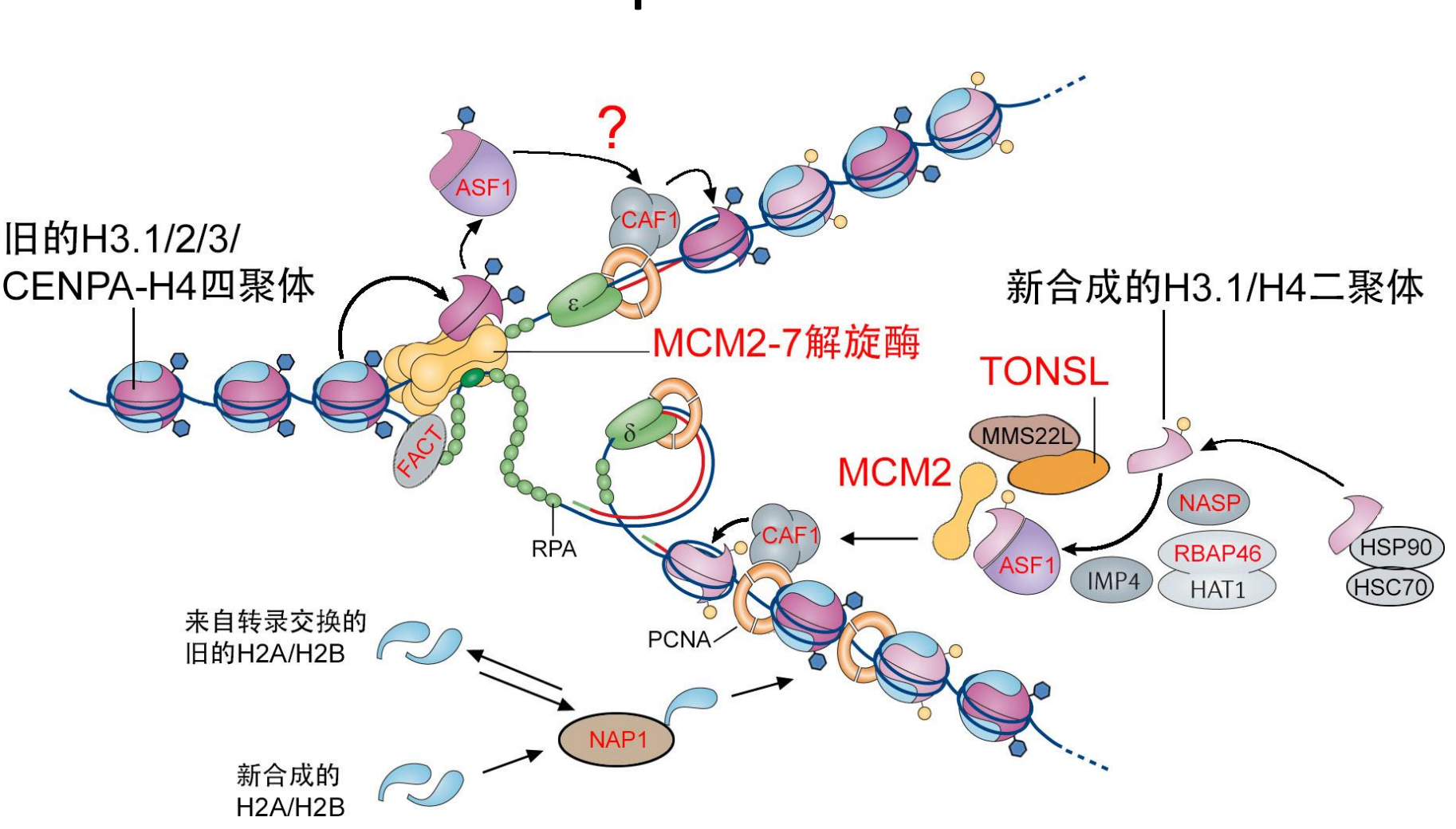
Nucleosomes Assembly
Nucleosomes Assembly happens during DNA Replication