L08 Epigenetics
一、Introduction to Epigenetics
Literally, epigenetics means above, or on top of, genetics.
Usually this means information coded beyond the DNA sequence, such as in covalent modifications to the DNA or modifications to the chromatin structure.
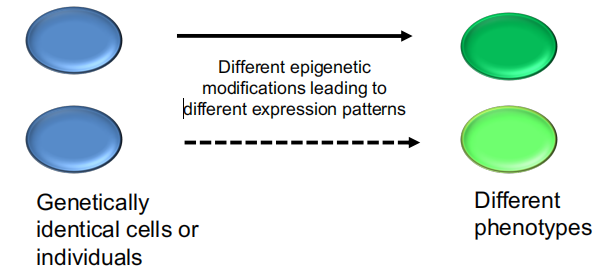
Practically, epigenetics describes phenomena in which genetically identical cells or organisms express their genomes differently, causing phenotypic differences.
The epigenetics programming in plants helps silence transposons and maintain centromere function, and helps control developmental transitions
Epigenetics in Plants
Epigenetic controls in whole-plant processes
Transposon silencing
Control of flowering time
Control of imprinted genes
Gene silencing in trans; paramutation
Resetting the epigenome
Epigenetics in Animals
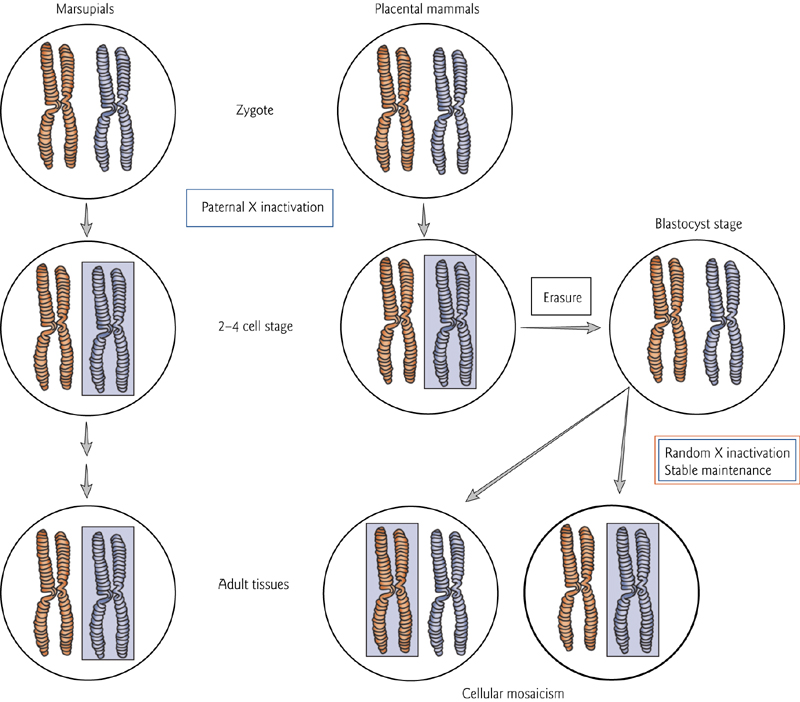
- X chromosome inactivation:
- In female mammals, one copy of the X chromosome in each cell is epigenetically inactivated.
- Paternal X-chromosome is preferentially inactivated. In mammals, this is erased in the blastocyst, but re-established in later stages…but it’s random which X-chromosome is inactivated.
二、Mechanism of Epigenetics
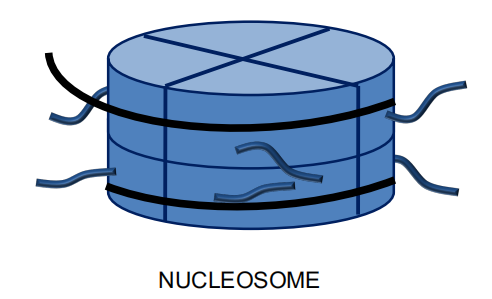
Nucleosomes
Approximately 147 base pairs of DNA wrapped around a histone octamer
Consists of 8 histones:
- 2 each: H2A, H2B, H3, H4

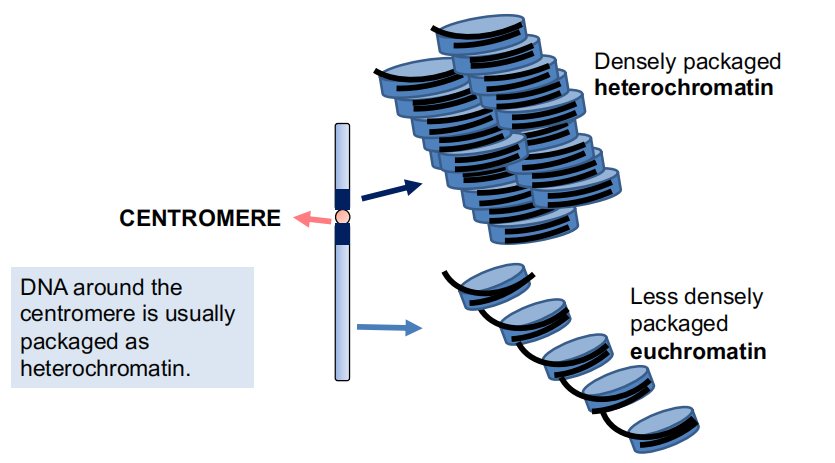
Heterochromatin and Euchromatin
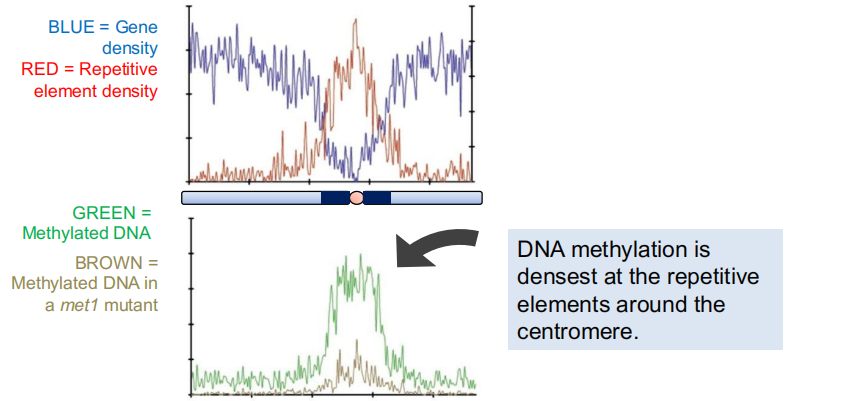
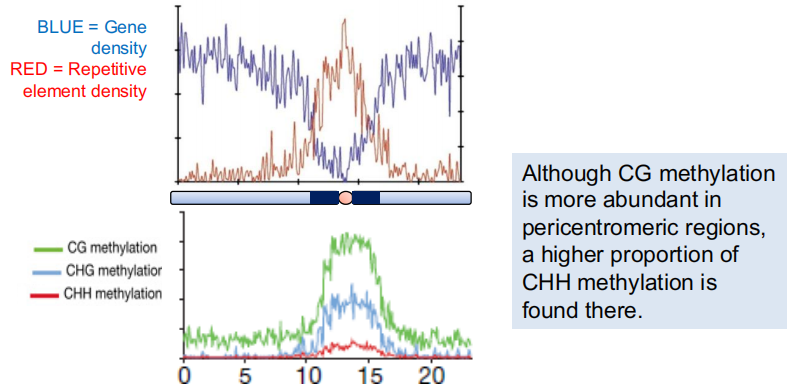
DNA around the centromere is usually packaged as heterochromatin.
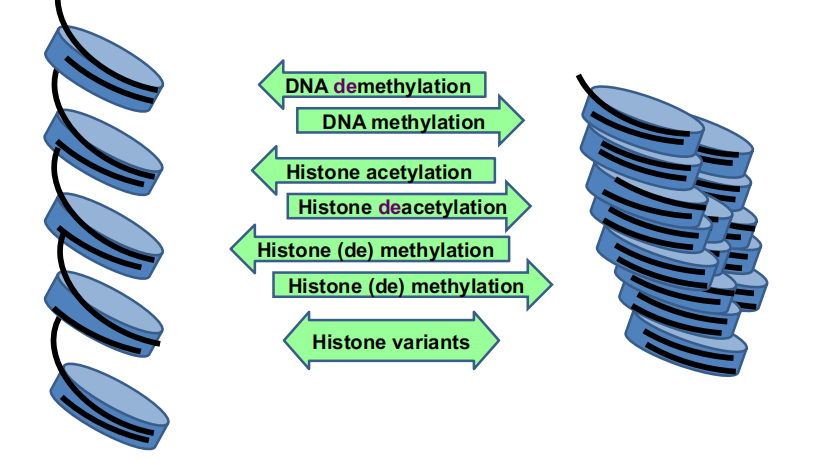
Epigenetic modifications
Epigenetic modifications include:
- Cytosine methylation of DNA
- Histone modifications
Collectively, these changes contribute to the distribution of DNA into silent, heterochromatin and active euchromatin
1. DNA Methylation
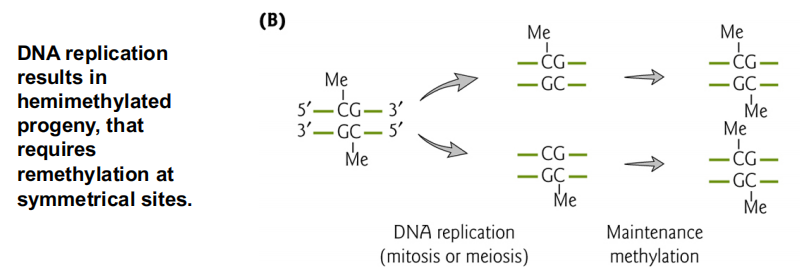
DNA methylation is an epigenetic mark maintained through replication

DNA can be covalently modified by cytosine methylation.
The methylation is a negative regulatory role
Different DNA methylases act on different sites
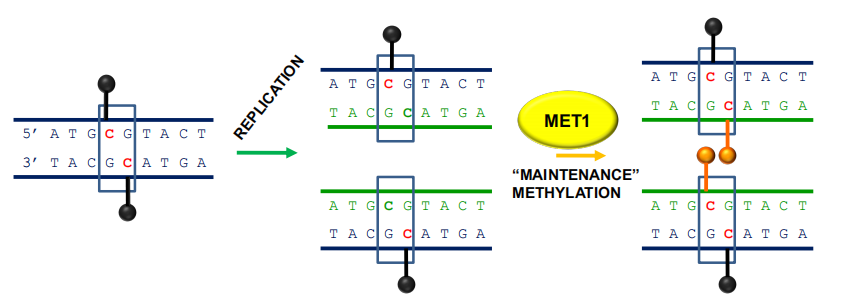
CG methylation can be propagated during DNA replication
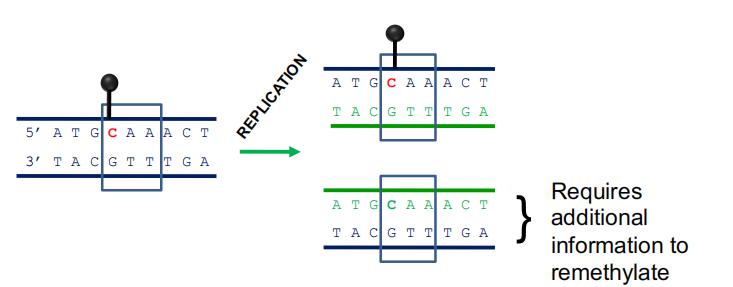
Asymmetric methylation sites require additional information
- Asymmetric methylation sites are maintained (and initiated) by information on associated histones, and an RNA-based mechanism, RNA-directed DNA Methylation (RdDM), that directs DNA methylases to these sites.
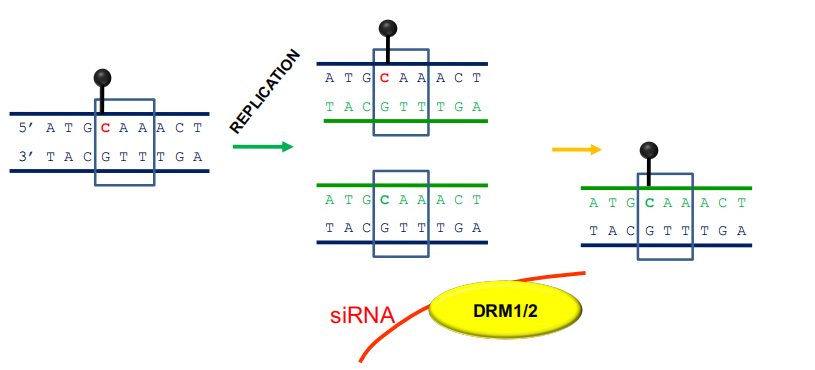
Some sites are maintained by small interfering RNAs (siRNAs)
DNA methyltransferases in Arabidopsis
- MET1 (METHYLTRANSFERASE1) – 5’-CG-3’ sites
- Silencing of transposons, repetitive elements, some imprinted gene
- CMT3 (CHROMOMETHYLASE3) – 5’-CHG-3’ sites
- (H= A, C or T)
- Interacts with histone marks/modifications
- DRM 1 & DRM 2 (DOMAINS REARRANGED 1 and 2) - 5’-CHH-3’ sites
- Primarily targets repetitive elements
- Requires the active targeting of siRNAs
Interference of DNA methyltransferase activity causes phenotypic alternations
Heterochromatin DNA is highly methylated
2. Histone Modification
Histone can be modified to affect chromatin structure
The amino terminal regions of the histone monomers extend beyond the nucleosome and are accessible for modification.
Different Histone Modifications are Associated with Genes And Transposons
Histone can be modified by:
- Acetylation (Ac,乙酰化)
- Ubiquitination (Ub,泛素化)
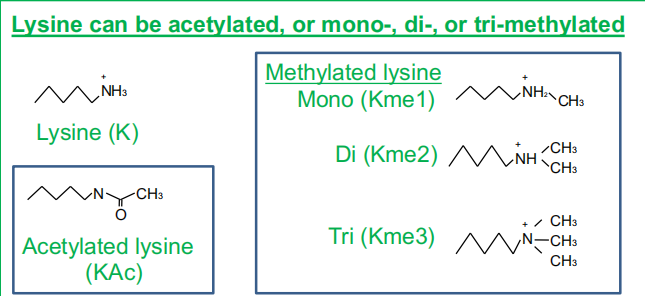
- Methylation (Me)
- Phosphorylation (P)
- Sumoylation (Su,糖基化)
Depending on their position, these can contribute to transcriptional activation or inactivation.
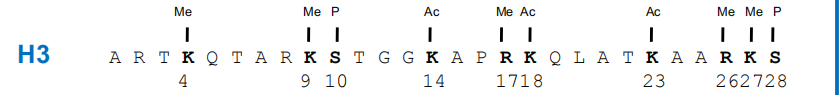
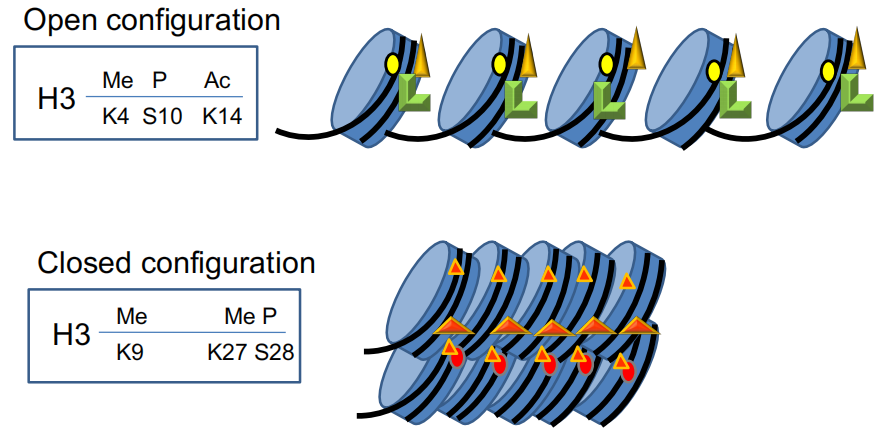
(1) Example – H3 Modifications
The amino terminus of H3 is often modified at one or more positions, which can contribute to an activation or inhibition of transcription.
Histone Modification affects the chromatin structure
Small RNAs
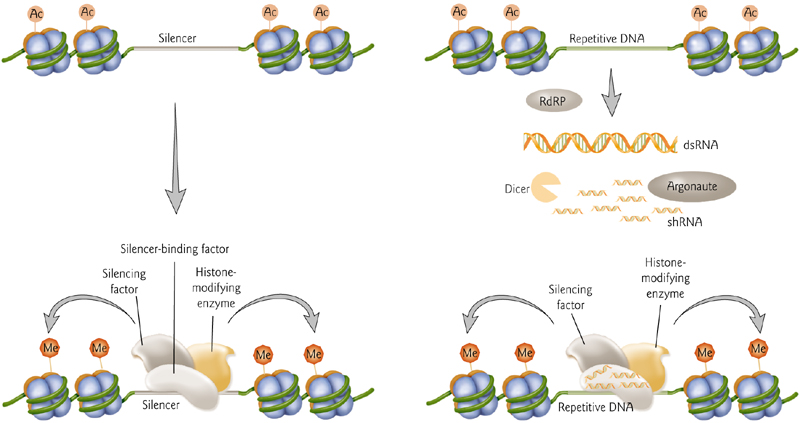
Small RNAs can direct Histone Modifications to form Heterochromatin
Repetitive sequences may be targeted for silencing by small RNAs produced from those repeats.
Regulators of Chromatin – Change in Binding Proteins
Different types of proteins recognize and bind modified histones, and these modifications can regulate the availability of the DNA to different activities.
3. Summary
三、Analysis of DNA Methylation
Compare digestion patterns using a pair of enzymes that are sensitive or not to DNA methylation.
- HpaII doesn’t cut at $CC^mGG$.
- MspI cuts at both $CC^mGG$ and $CC\cdot GG$
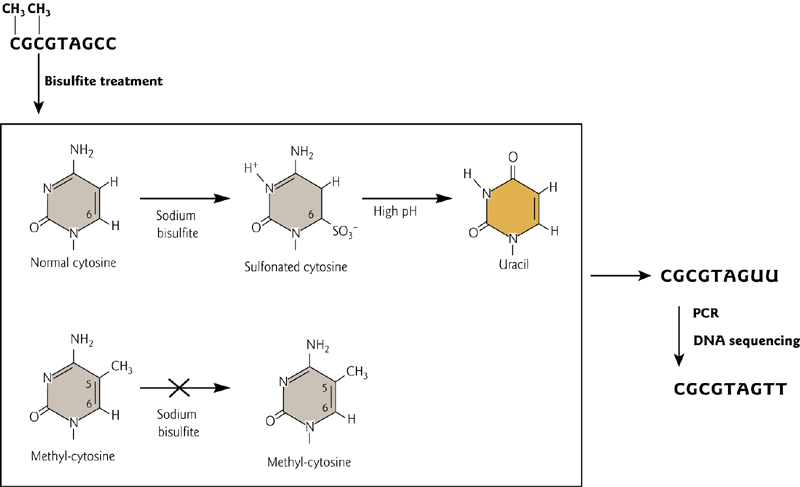
Sodium Bisulfite
Sodium Bisulfite treatment to detect methylated DNA
DNA methylation can be detected by treating DNA with sodium bisulfite to convert methylated cytosines to uracil, which becomes thymine after a PCR amplification step.
ChIP-SEQ (Chromatin Immunoprecipitation)
ChIP-SEQ allows the identification and analysis of histones with a wide variety of specific modifications, based on the DNA that they are associated with.
Epigenetic controls in whole-plant processes
- Transposon silencing
- Control of flowering time
- Control of imprinted genes
- Gene silencing in trans; paramutation
- Resetting the epigenome
X-Chromosome Inactiation
Paternal X-chromosome is preferentially inactivated.
In mammals, this is erased in the blastocyst, but re-established in later stages…but it’s random which X-chromosome is inactivated.