L11 Cloning Vectors and Host cells
一、Basic features of Cloning Vectors.
Why the Vector and the Host Cell are important ?
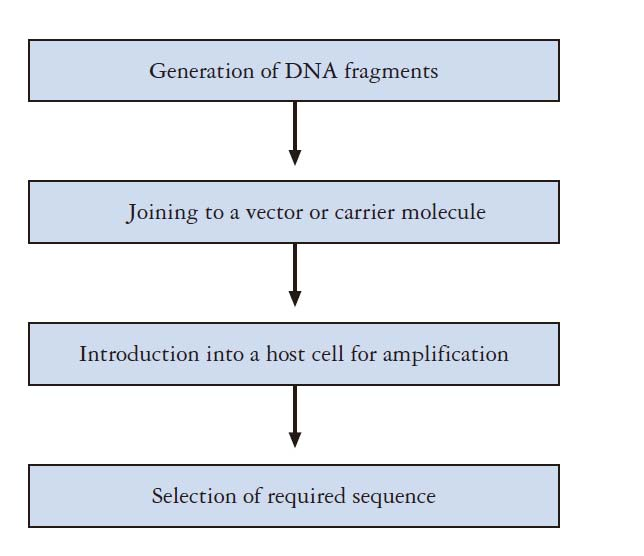
- They enables manipulation of genetic materials in living cells.
- A particular gene can be isolated and its nucleotide sequence determined and analyzed.
- Protein/enzyme/RNA function can be investigated.
- Mutations can be identified, e.g. gene defects related to specific diseases.
- Target protein or RNA can be expressed and purified.
What would be ideal features for a cloning vector ?
It should be able to replicate autonomously in the host cell.
It should be of a reasonable size, facilitating the cloning procedure
Harbors sequence/modules for cloning
Easy to be isolated & purified.
Easy to be screened and identified in the host cell.
Basic features of (Cloning) Vectors
Modules for replication: Origin of replication – Different Origin largely determined the copy number.
Modules for insert/cut-out the gene of interest: Multiple Cloning sites or recombination site
Modules for select the transformants: Antibiotic resistance genes; Reporter genes.
Modules for the control of target gene: promoter, operator, ribosome binding site. (for expression vector only)
TYPES OF VECTORS
| Vectors | Targeted Host |
|---|---|
| Plasmid | Bacteria, Streptomyces |
| Phages Vector | Bacteria |
| Cosmid | Bacteria |
| Yeast Cloning Vectors | Yeasts |
| Human Artificial Chromosome | Human Cell line |
| SV40 vector | Mammalian Cell |
二、A general introduction about plasmid
Plasmid vector
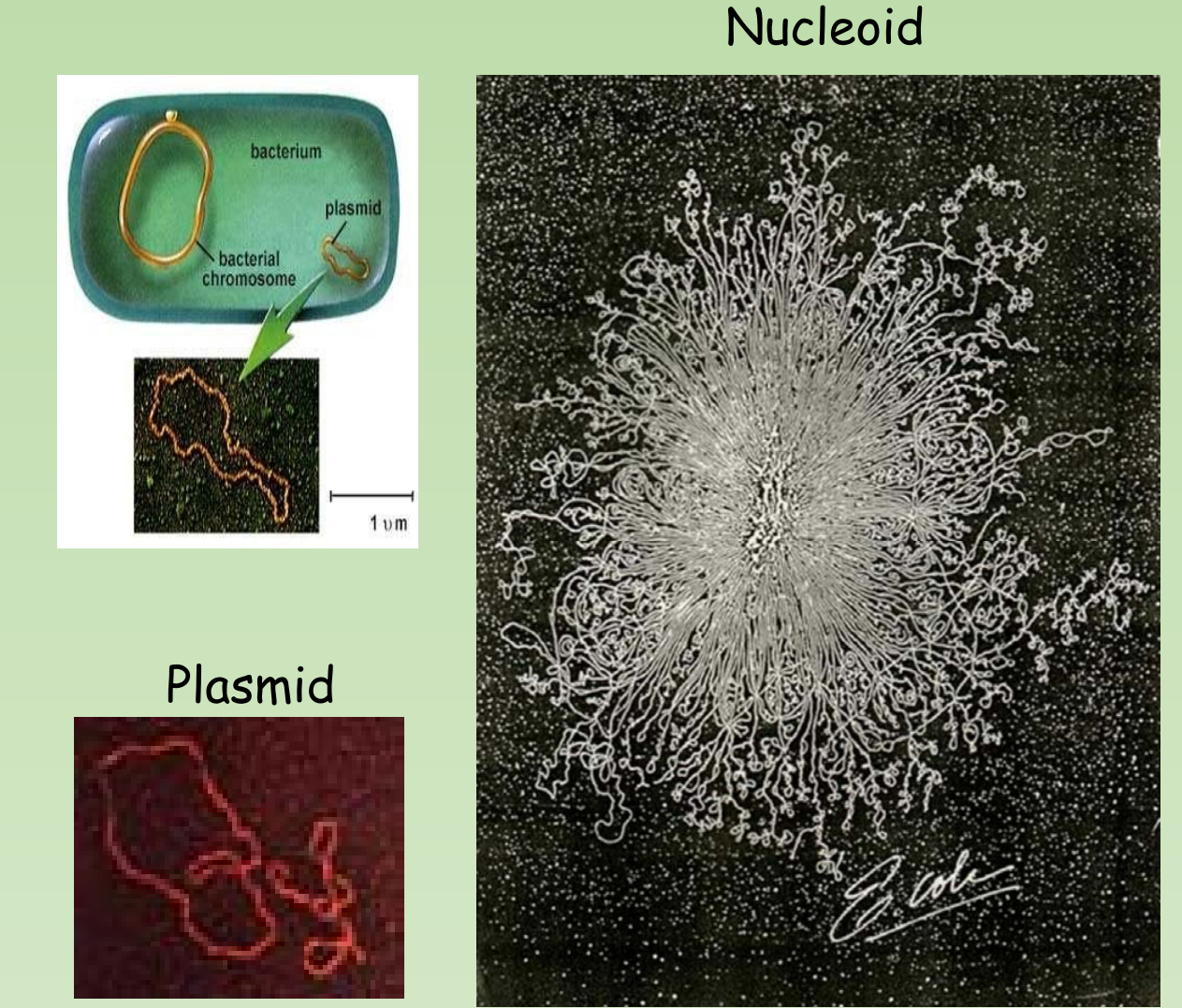
- Plasmid vector is a small piece of circular DNA found outside of the bacterial chromosome.
- Capable of autonomous replication
- Can transfer genes from one cell to other.
1. Features of Plasmid
Small (Versus host genome), Extrachromosomal DNA, originally found as naturally occurring.
Normally dispensable, but can have selective advantage under certain condition.
Plasmid could mediate their own transfer between Bacteria.
- Conjugative vs Non-conjugative, Mobilization
Must be a replicon, must replicate along with host cell division, otherwise will be lost
High copy plasmids are usually small; low copy plasmids can be large
Replication is strictly controlled for low copy plasmid.
- Stringent control vs Relaxed control
Plasmid replication requires host cell functions.
Copy number is regulated by initiation of plasmid replication.
Stringent control vs Relaxed control
2. Varieties Of Plasmids Based On Functions
(based on function there are five main classes:)
- Fertility / F- plasmids: They are capable of conjugation and result in the expression of sex pilli.
- Resistance (R) plasmids: They contain genes that provide resistance against antibiotics or poisons.
- Col plasmids: They contain genes that code for bacteriocins, proteins that can kill other bacteria.
- Degradative plasmids: They enable the digestion of unusual substances, e.g. toluene and salicylic acid.
- Virulence plasmids: They turn the bacterium into a pathogen
Typical features of a cloning plasmid: learn from pUC18
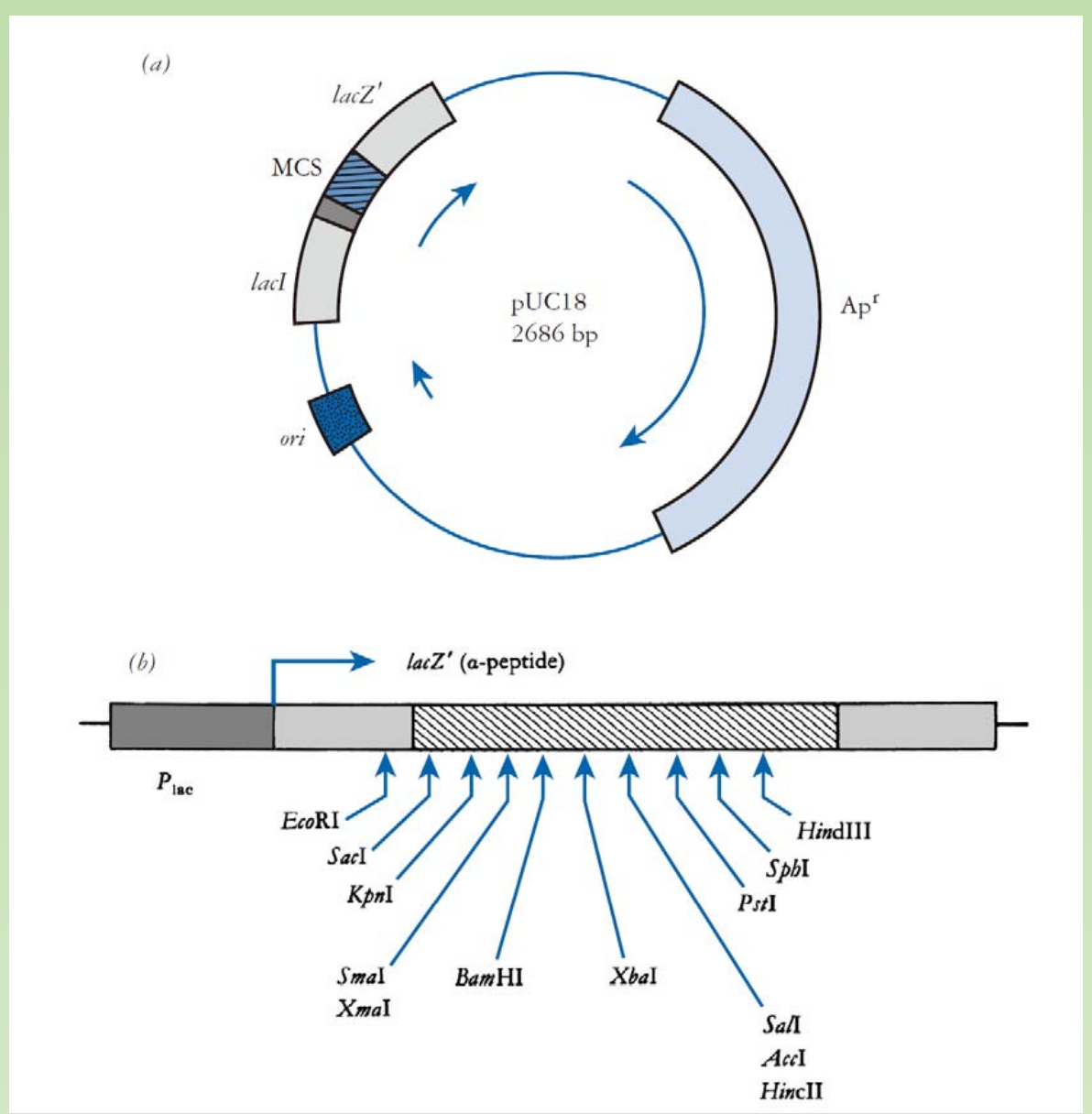
A classical cloning vector
- Small size
- Origin of replication
- Multiple cloning site (MCS)
- Selectable marker genes
pUC18 is one of a series of plasmid cloning vectors created by Joachim Messing and co-workers.
Nomenclature:
- p =plasmid
- UC = University of California
- 18 = a number given to distinguish it from others
pBR322 = $\text{plasmid}^{\text{Boliver Rodriguez}}$
Antibiotic resistance gene: a gene for antibiotic resistance to Ampicillin (ampR).
- How does Ampicillin resistance works ?
A reporter gene (and its promoter) encode enzyme beta-galactosidase (lacZ).
- What’s the function of this reporter gene ?
Polylinker region (multipe cloning sites, MCS), with a series of unique restriction sites
found nowhere else in the plasmid.- Enabling cut and paste of your target DNA – You all know already !
Replication origin: enabling the replication within E.coli host cell.
Antibiotic resistance genes (ampicillin resistance & satellite colony)
1. Antibiotics
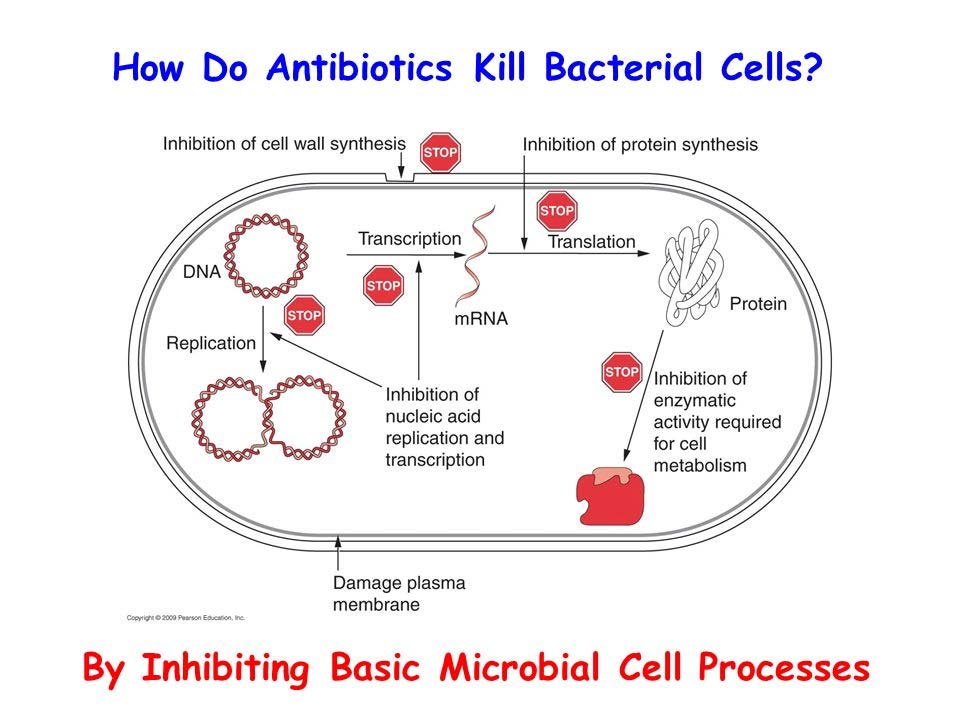
The antibiotics acts by inhibition of:
- Cell wall synthesis (e.g. Ampicillin)
- Translation
- Transcription
- DNA replication
- Other basal metabolic processes.
Different antibiotics:
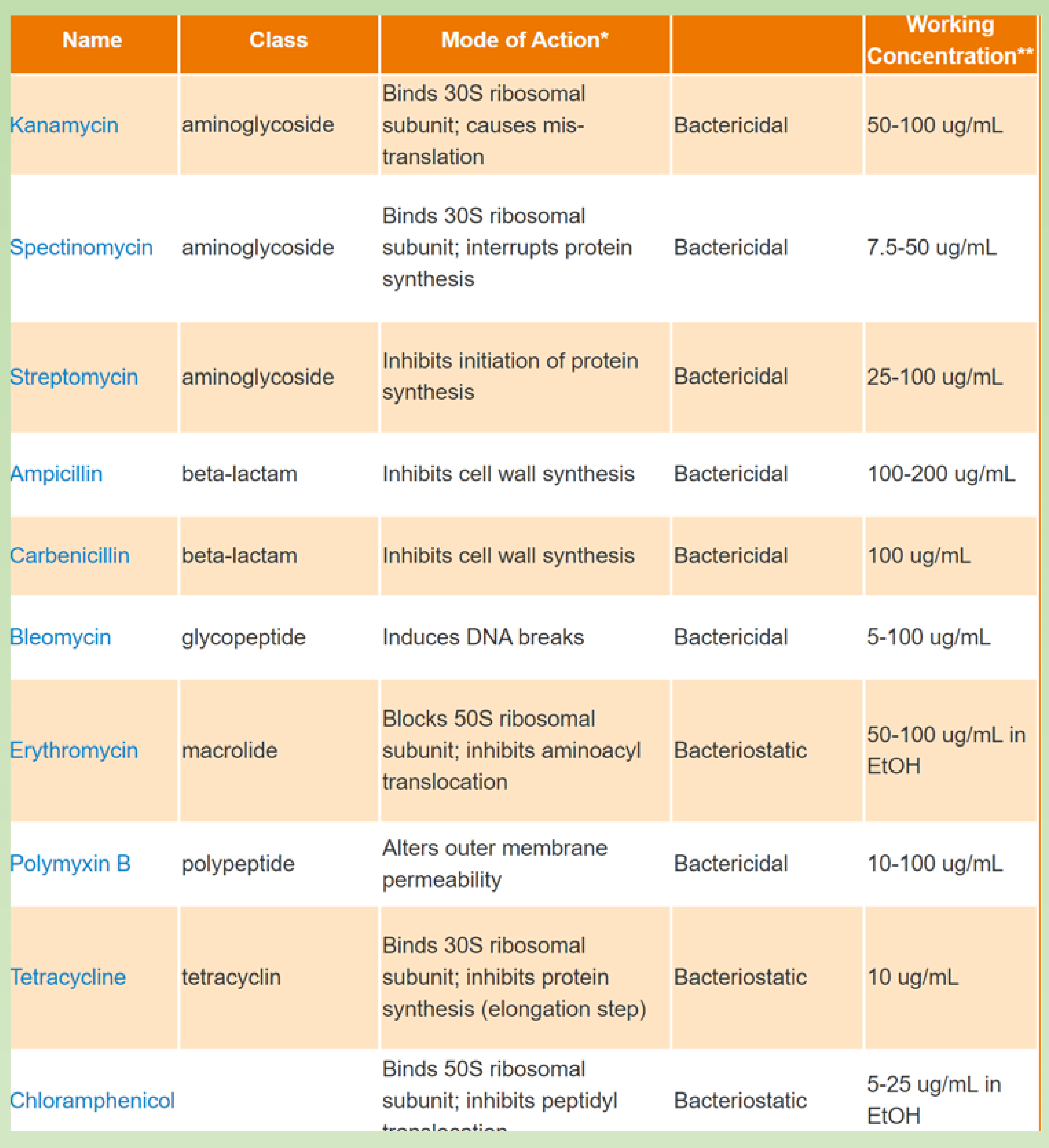
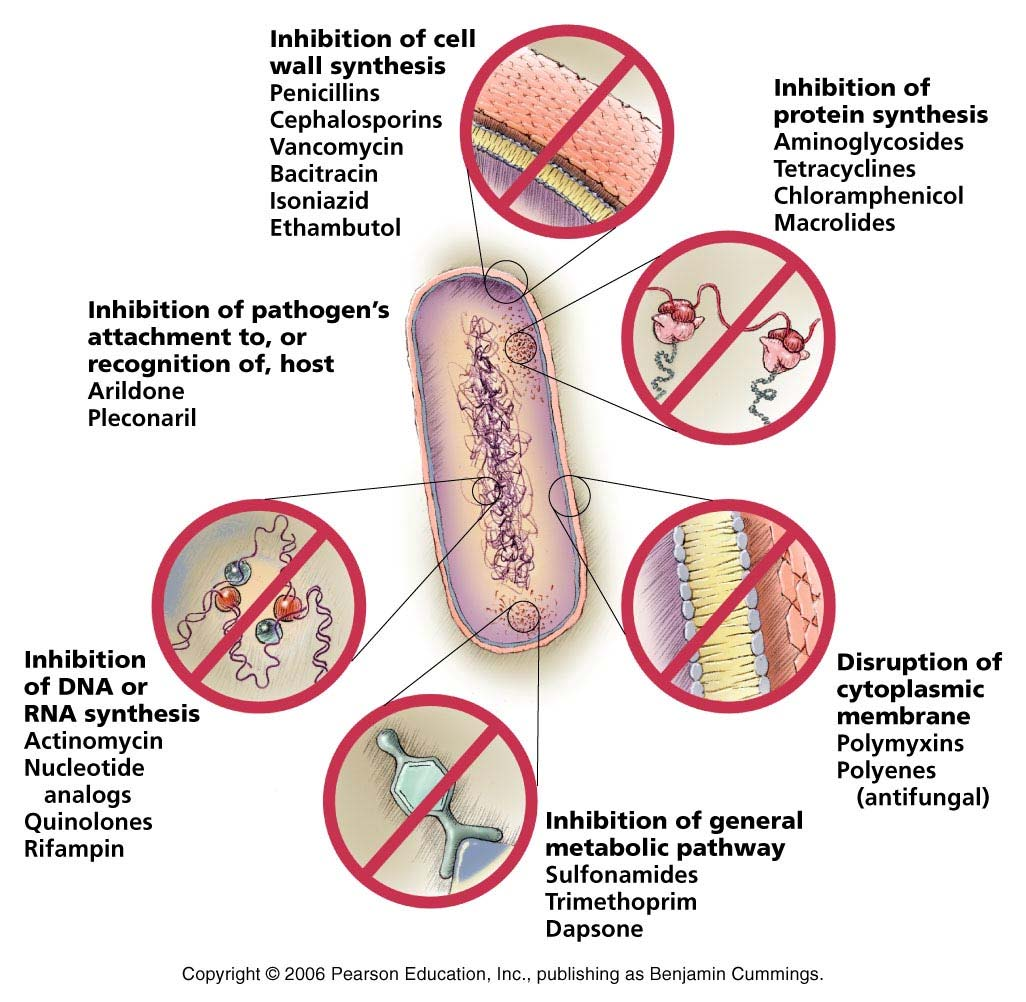
How does Ampicillin resistance works ?
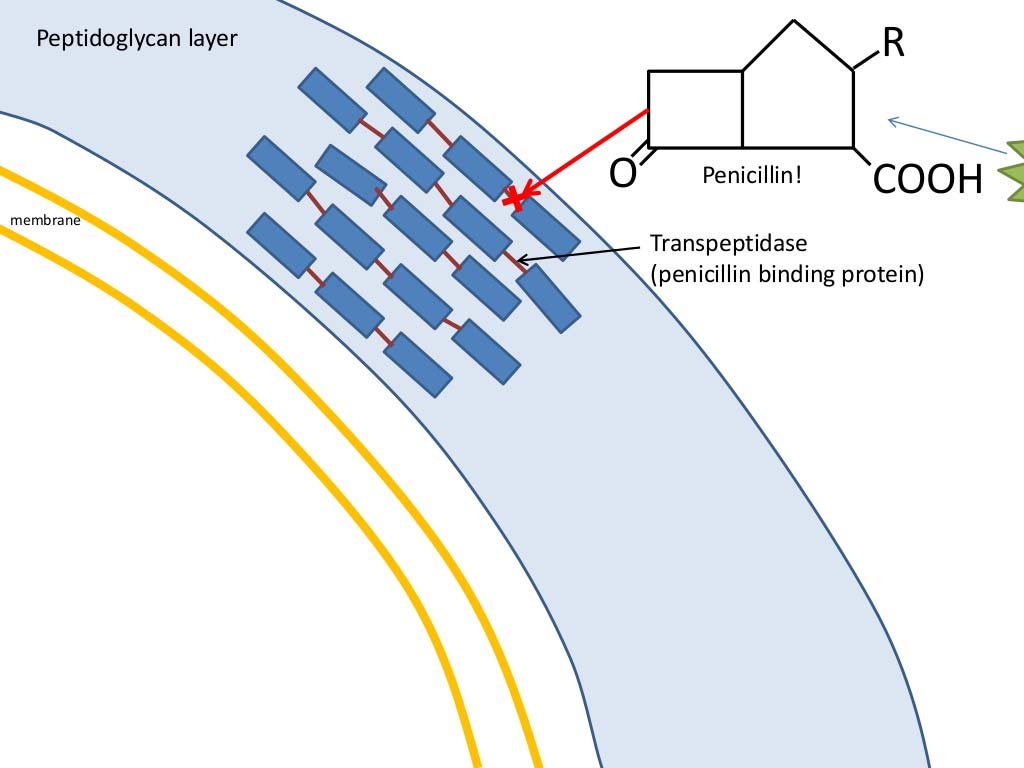
- Penicillin irreversibly blocks bacterial cell wall synthes is by inhibiting the formation of peptidoglycan cross-links. Penicillin covalently binds to the enzyme transpeptidase that links the peptidoglycan molecules in bacteria, it inhibits the molecule so that it cannot react any furt her and cell wall cannot be further synthesized.
2. AmpR: β-Lactamases
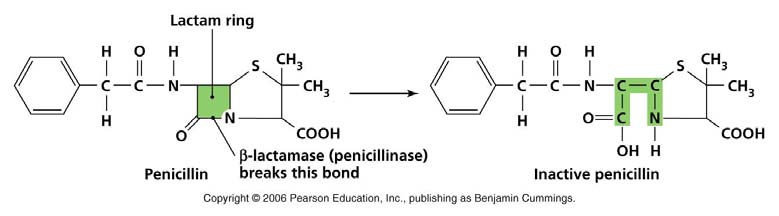
- Ampicillin is a derivatives of penicillin.
- The ampicillin resistance gene (AmpR) encode beta-lactamase.Through hydrolysis, the enzyme lactamase breaks the β-lactam ring open, deactivating the molecule’s antibacterial properties
- Penicillin (or Ampicillin) kills bacteria by interfering with the ability to synthesize cell wall
- In the presence of penicillin, bacteria cell often ruptures during cell division.
Ampicillin and the Satellite Colonies ?
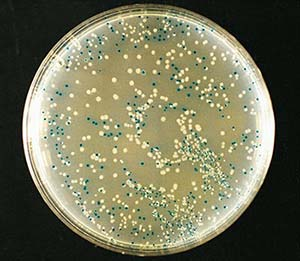
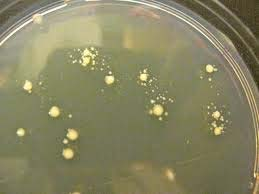
- Satellite colonies are very small colonies of cells that have not taken up the plasmid that form around a large colony that has taken up the plasmid. The satellites form because the beta -lactamase released by the AmpR expressing colony degrades the ampicillin in the vicinity of the colony.
- Would satellite colonies forming after you transform a Kanamycin containing plasmid ?
- The answer is no
LacZ reporter genes (blue & write selection)
1. beta-galactosidase (lacZ) & Blue and Write selection
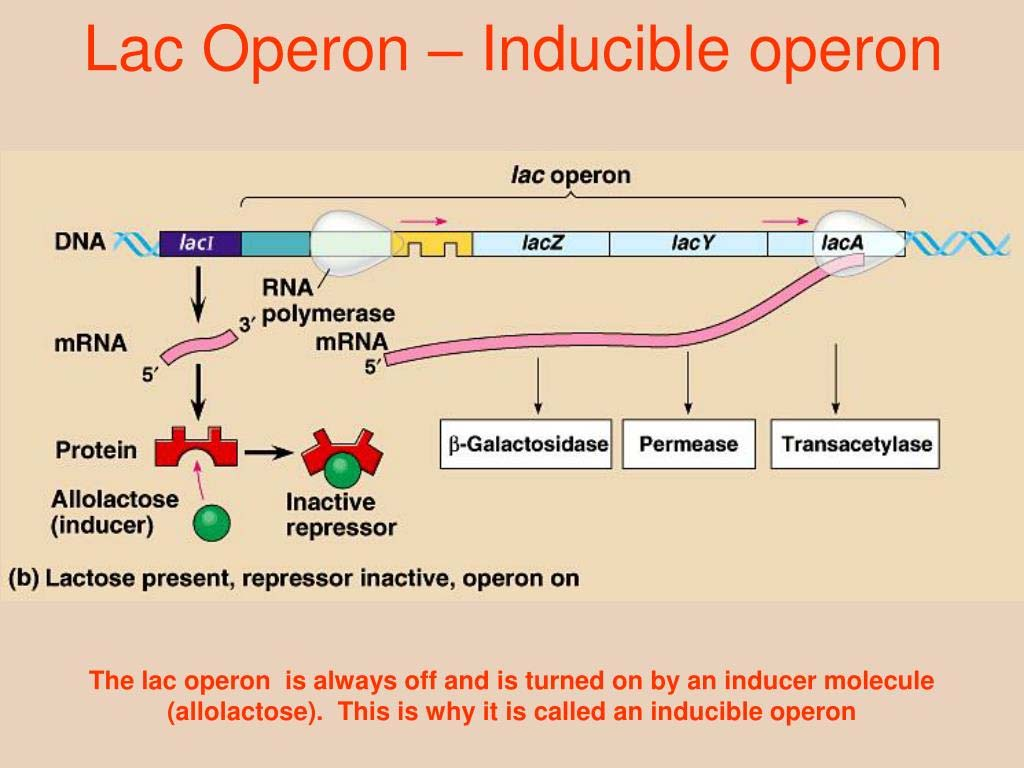
- Suppressor protein (LacI ) bound to the operator sequence to block the engagement of RNA polymerase.
- When lactose come, (allolactose), it binds to the LacI protein, triggers an conformational change and lacI no longer able to block the polymerase
- LacZ, which encode the beta-glactosidase is now able to express and mediate the hydrolysis of lactose into glactose and glucose.
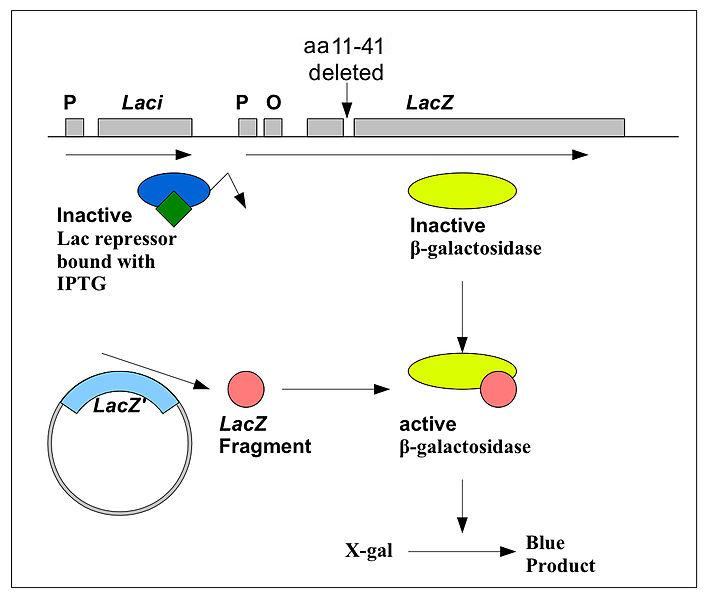
Three key words
- alpha complementation.
- It turned out that the Lac Z gene in the E.coli for the cloning is an truncated version, a 30 amino acids long piece of it is missing
- This 30 aa piece is actually in the plasmid you are going to use. The 30 aa piece + the rest of the LacZ can then perform as the natural ones.
- The 30aa call alpha peptide, and this reaction called alpha complementation.
- IPTG
- IPTG and X-Gel: In fact, we use the IPTG to inactivate LacI. IPTG is a structural mimic of allolactose. And X-gal as substrate of LacZ. If alpha complementation happened successfully, X-gal will be hydrolyzed and part of it becomes a blue precipitate.
- If there is insert in the alpha peptide, the colony would looks white.
- X-gal
IPTG & X-Gal
- IPTG is a structural mimic of allolactose. And X-gal as substrate of LacZ.
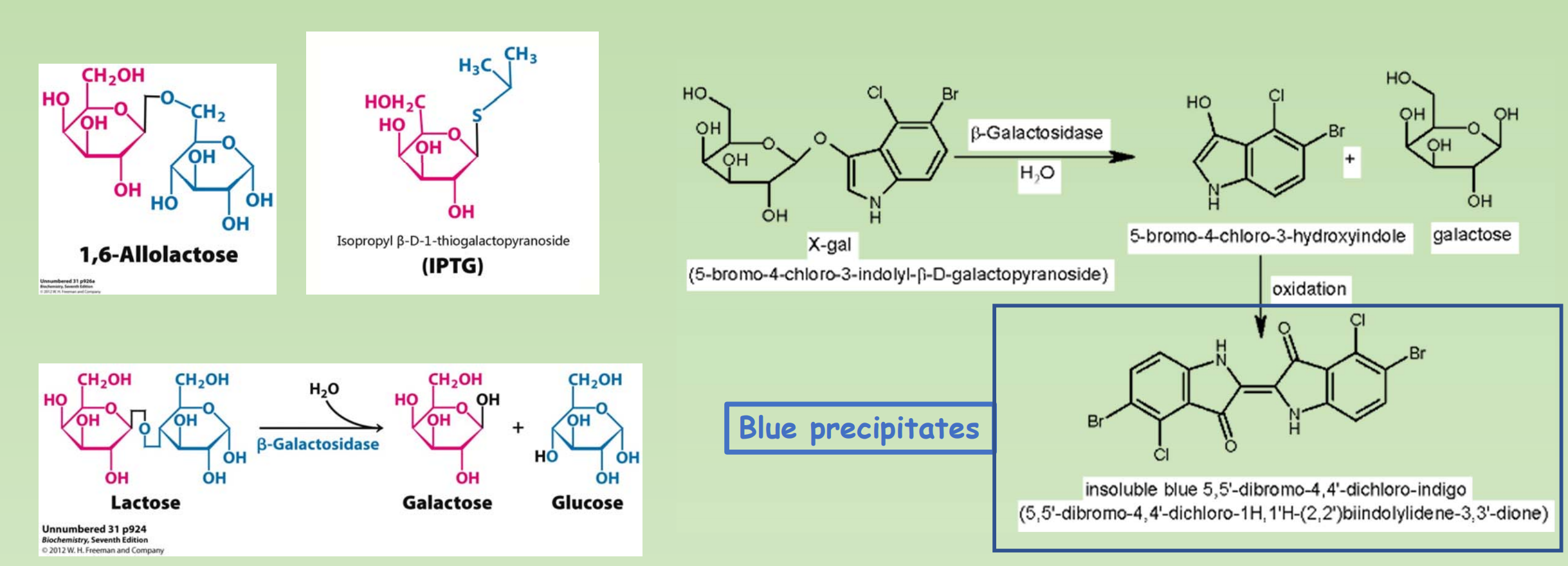
- IPTG: Isopropyl β-D-Thiogalactoside (异丙基硫代半乳糖苷)
- X-gal: 5-Bromo-4-chloro-3-indolyl β-D-galactopyranoside ( 5-溴-4-氯-3-吲哚-β-D-半乳糖苷)
2. Look at pUC18 again

- LacZ: beta-galactosidase
- Alpha peptide
- lacZΔM15 deletion mutation.
- Alpha complementation
- MCS (multiple cloning site)
- IPTG
- X-gal
The Multiple cloning site located within the alpha peptide coding region, so an insertion in this region will disrupt the alpha peptide.
3. Other features that might exist in a cloning plasmid
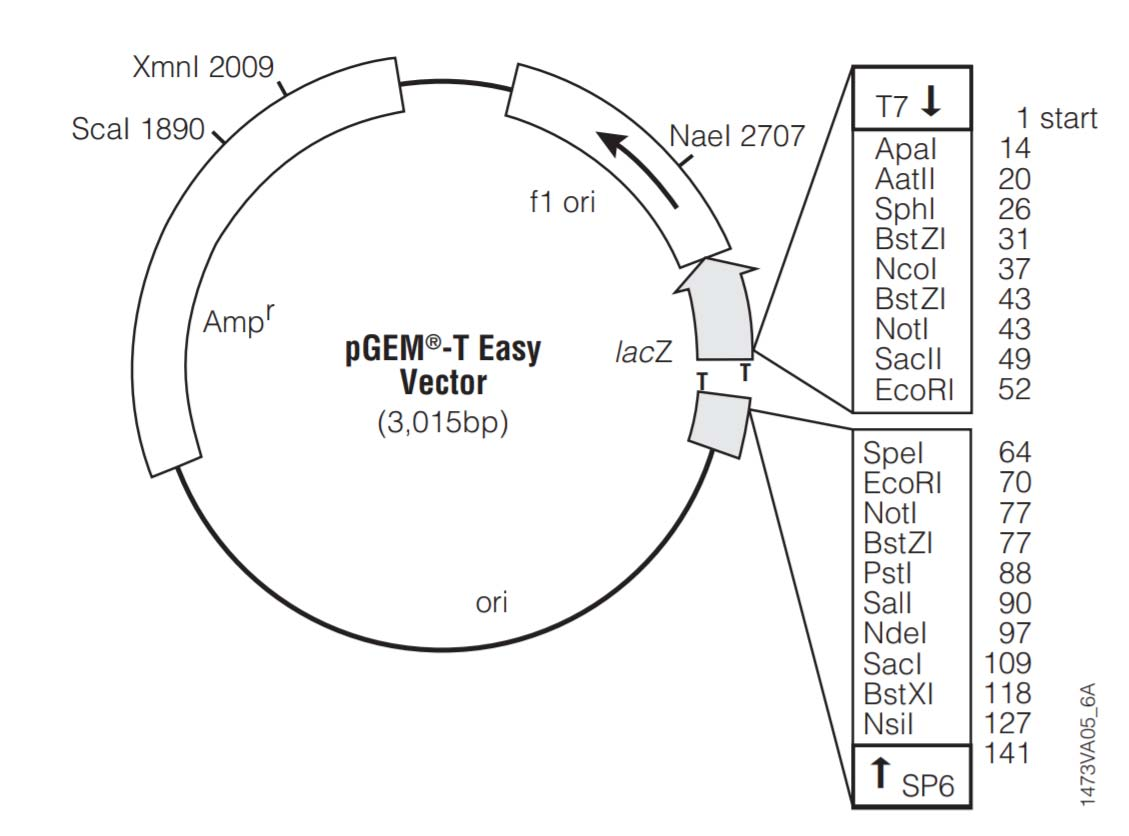
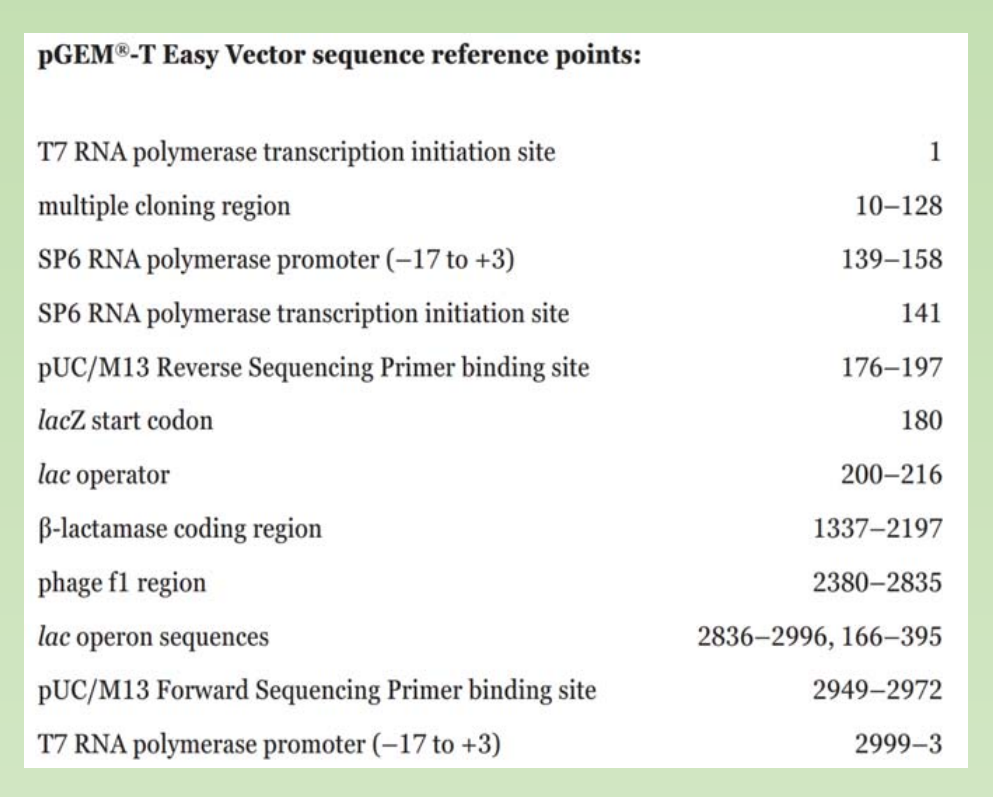
- Sequencing primer binding site
- Phage promoter for RNA synthesis. (T7, SP6, T3).
三、Plasmid conformation and its purification
The conformation of a plasmid
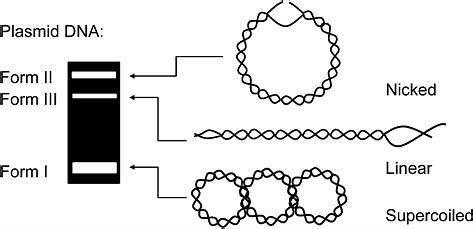
- How many conformation does a plasmid has
- How many bands would you expect when you run your extracted plasmid on gel ?
In General, a plasmid have three different conformations,
- Nicked
- Linear
- Supercoiled
Supercoiled DNA:

DNA supercoiling refers to the over or under-winding of a DNA strand, and is an expression of the strain on that strand. Supercoiling is important in a number of biological processes, such as compacting DNA, and by regulating access to the genetic code, DNA supercoiling strongly affects DNA metabolism and possibly gene expression. Additionally, certain enzymes such as topoisomerases are able to change DNA topology to facilitate functions such as DNA replication or transcription.
How many bands would you expect when you run your extracted plasmid on gel ?
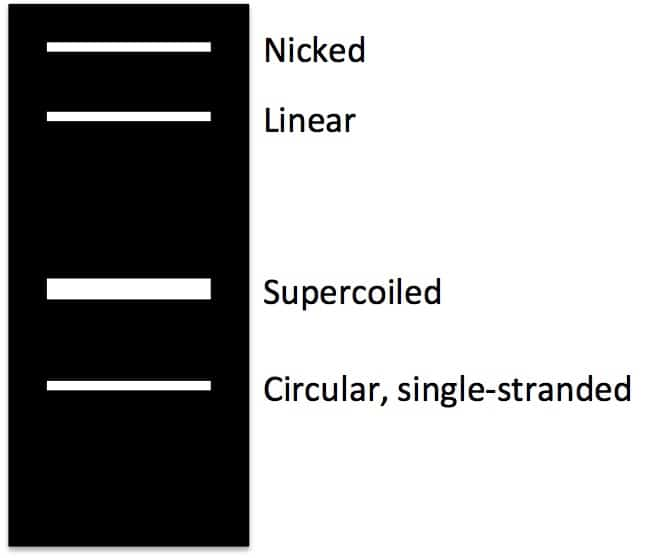
- You are likely to observe 4 bands when you resolve your plasmid on gel.
- Single stranded circular DNA (sscDNA)
- Supercoiled
- Linear
- Nicked
The ss-circular DNA exist likely due to the incomplete reannealing of the two stands of dsDNA during the plasmid mini-prep
Plasmid Purification: Plasmid purification through CsCl gradient centrifugation
“ Old School method of purifying plasmid”:
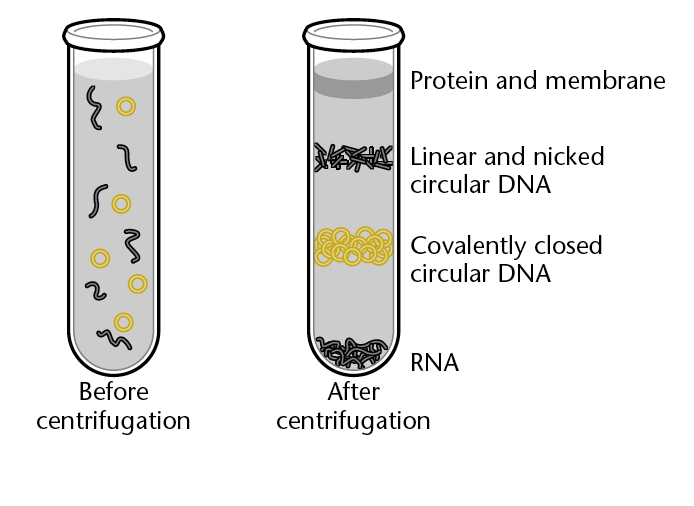
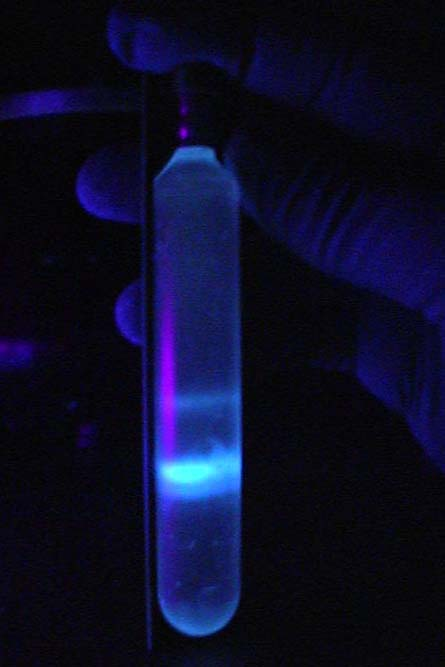
- CsCl gradient with ethidium bromide and UV light.
1. Plasmid Purification: Alkaline lysis method of plasmid purification

What are the components in P1, P2 and N3 ?
Solution I: 25mM Tris-HCl, pH8.0, 10mM EDTA, (50mM glucose in some protocol) 100ul – optional lysozyme treatment when dealing with gram positive strain. Lysozyme digest cell wall made of peptidoglycan.
Solution II: 200mM NaOH, 1% SDS, 200ul. At high pH, then, the solution is rich in hydroxide ions, and these negatively-charged ions can pull hydrogen ions off of molecules like the base pairs in DNA. This process disrupts the hydrogen bonding that holds the two DNA strands together, causing them to separate.
Reminder: Alkaline hydrolysis and Alkaline denaturation
Solution III: 3 M potassium acetate, pH 4.8, 150ul
- After adding the solution, the solution is neutralized suddenly. The DNA in a plasmid have good chance to re-anneal between two strand. The genomic DNA is too large to be re-annealed, so form precipitate
The alkaline lysis can be modified to be used together with the column. Include the chaotropic salt into the solutions help with the binding to the column.
Transformation & Transduction & Transfection
Transformation – put a plasmid into host cell
- Heat shock 30min - 90s – 5mins, create tiny holes in the membrane to allow for efficient transformations
- Competent Cell – CaCl2 (0.1M) most classical method (E.coli grow to the pre-log phase) OD600 = 0.2-0.6.
- Super-competent Cell – grew cell in low temperature + MnCl2 been included in the wash buffer. You can try to make your own.
- Transformation efficiency, 10^6 to 10^9 clones per 1ug of DNA, very low.
Colony-forming unit, or CFU & Plaque-forming unit, or PFU
Transduction - is the process of transferring any bacterial gene to a second bacterium through a bacteriophage
Transfection – transfer the DNA into cell through virus (phage)
Transfection is the process of deliberately introducing naked or purified nucleic acids into eukaryotic cells. Refers specifically virus infection based methods
四、A very brief introduction about E.coli.
Introduction and history
A brief introduction of the E.coli stains that we used in Molecular Cloning
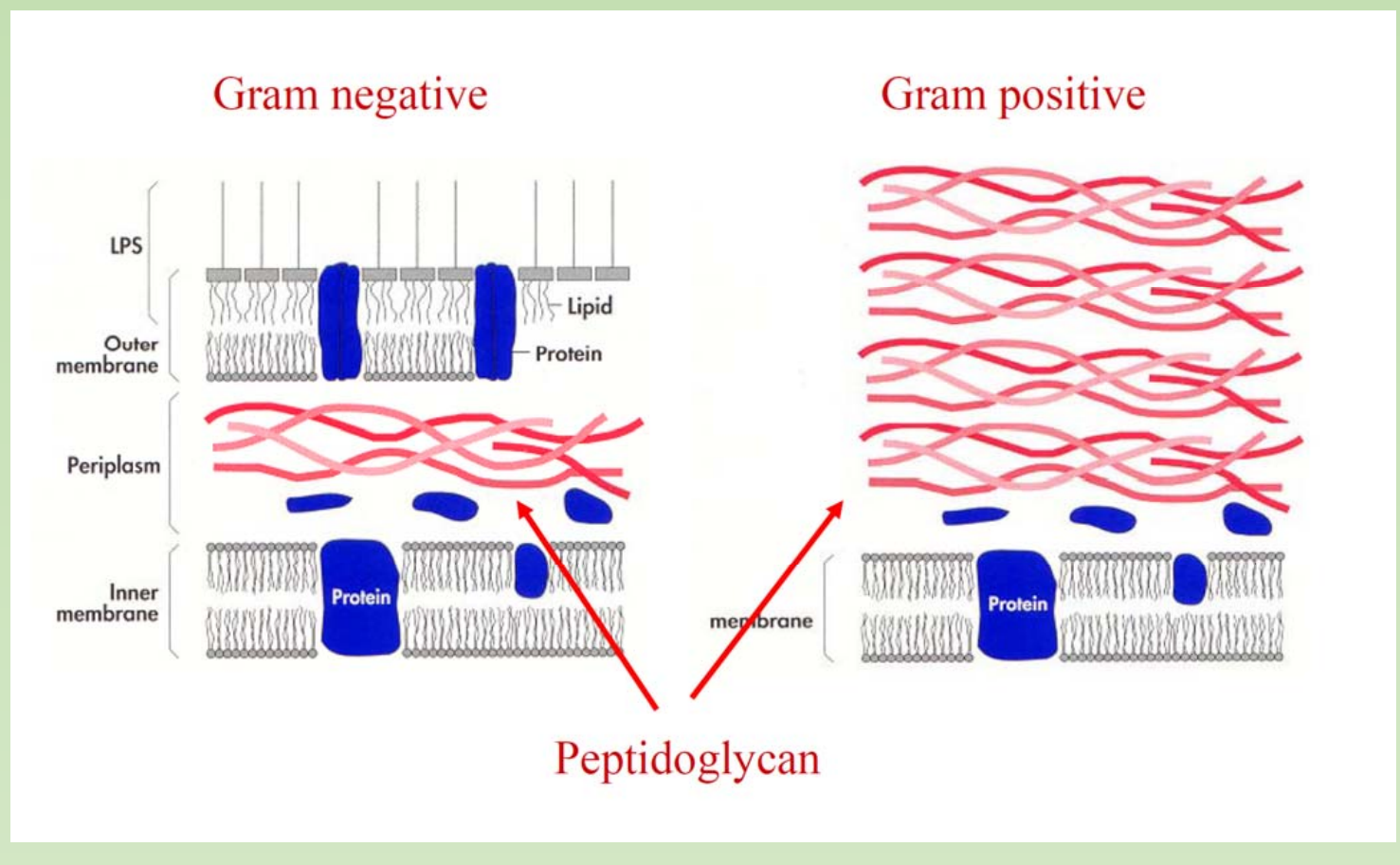
What would be a ideal host cell?
- The host cell should be compatible with the features of cloning plasmid that we uses
E. coli are gram-negative
- Would it be necessary to do a lysozyme treatment when we extract plasmid from E.coli ?
- the answer is no, the thick layer of peptidoglycan of gram positive strain would require lysozyme treatment.
E. coli named after Dr. Theodor Escherich, who first described it back to 1885. There are quite some strains, some of which are deadly to humans
All of currently used strain in genetic engineering derived from K-12 strain – DH5alpha, DH10b (Top10), DB3.1 and the B strain. – BL21, Rosetta , they are not pathogen.
- Currently used strains have all kinds of mutations.
DH5alpha:
- F-, endA1, glnV44, thi-1, recA1, relA1, gyrA96, deoR, nupG, Φ80d, lacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK- mK+), λ–
A few common mutations
F-, endA1, glnV44, thi-1, recA1, relA1, gyrA96, deoR, nupG, Φ80d lacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK- mK+), λ–
Mutation enables blue/write screening. lacZ
- Φ80d lacZΔM15, delete a small fragment in LacZ, enables alpha complementation.
Mutation leads to Nutrient Deficient
- cannot growth in nature environment (thi-1: This strain requires thiamine (thiamine auxotroph, cannot produce its own thiamine))
Mutation to stabilize the Plasmid,
- Reduce chance of recombination. (Mutation in a DNA-dependent ATPase that is essential for recombination and general DNA repair, recA1)
- Increase yield.
- Endonuclease A deficient, endA1
- deoR: The transcriptional repressor DeoR, for “Deoxyribose Regulator”. Allow produce the decoxyribose constantly, therefore can take large plasmid.
- nupG: NupG is one of two high-affinity nucleoside transporters in E. coli.
- Inactivate endogenous restriction/modification systems
- hsdR17(rK- mK+), mutation eliminate restriction enzyme Ecok1,
- DNA without mEcok1 modification won’t be cut.
- Ecok1-mEcok1 (restriction-modification system)
There are lots of different strains
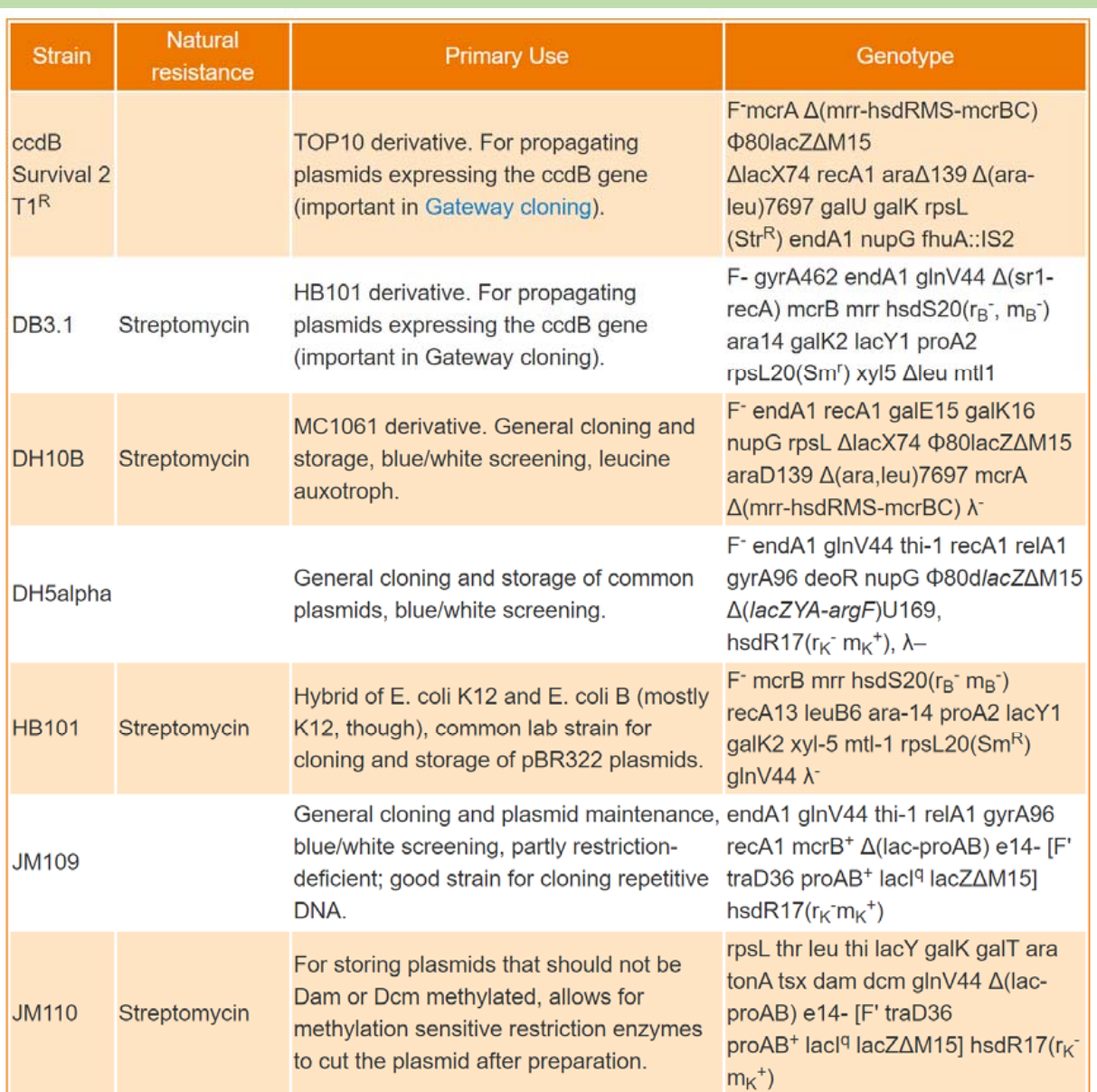
Most commonly used strains
1. Strains
K-12 strains
- DH5alpha, - general cloning
- DH10b (Top10) – general cloning
- DB3.1 - Gateway cloning, to propagate any plasmid that contains ccdB gene.
B strains
- BL21, - recombinant protein expression
- Rosetta - recombinant protein expression with rare codon optimization.
2. BL21 and Rosetta Strains for recombinant protein expression
BL21(DE3) pLysS
E. coli str. B F– ompT gal dcm lon hsdSB(rB–m B–) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λS) pLysS[*T7p20 ori*p15A](CmR)
- Deficient in Lon and OmpT proteases
- an E. coli B strain with DE3, a λ prophage carrying the T7 RNA polymerase gene and lacI
- Transformed plasmids containing T7 promoter driven expression are repressed until IPTG induction of T7 RNA polymerase from a lac promoter.
- pLysS plasmid chloramphenicol resistant; grow with chloramphenicol to retain plasmid
- The pLysS plasmid encodes T7 phage lysozyme, an inhibitor for T7 polymerase which reduces and almost eliminates expression from transformed T7 promoter containing plasmids when not induced.
Rosetta™(DE3)pLysS
E. coli str. B F– ompT gal dcm lon? hsdS**B(r**B–m**B–) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λS) pLysSRARE[*T7p20 ileX argU thrU tyrU glyT thrT argW metT leuW proL ori*p15A](CmR)
- an E. coli B strain with DE3, a λ prophage carrying the T7 RNA polymerase gene and lacIq
- Transformed plasmids containing T7 promoter driven expression are repressed until IPTG induction of T7 RNA polymerase from a lac promoter.
- Chloramphenicol resistant
- pLysSRARE contains tRNA genes argU, argW, ileX, glyT, leuW, proL, metT, thrT, tyrU, and thrU. The rare codons AGG, AGA, AUA, CUA, CCC, and GGA are supplemented
- The pLysS plasmid encodes T7 phage lysozyme, an inhibitor for T7 polymerase which reduces and almost eliminates expression from transformed T7 promoter containing plasmids when not induced
ccdB gene represent another useful selection systems in gene cloning
This is part of the toxin-antitoxin system that encoded by the ccd operon which located on the F sex factor plasmid of E. Coli.
- There are two genes, ccdA and ccdB.
- ccdB codes for a toxic protein that act on DNA gyrase, can ultimately kill the cell.
- ccdA codes for the antitoxin protein that protects the cell against the toxic CcdB.
Cells that lose ccdA through the loss of the F plasmid will be killed after cell division
DB3.1, which contains a mutant version of DNA gyrase (gyrA462) that is resistant to the toxic effects of CcdB
Cells with F- genotype is sensitive to the ccdB. Such as DH5alpha. Cells with F+ genotype is tolerant to ccdB. Such as JM109
Advantages & Disadvantages of Plasmid as vector
Advantages:
- Readily isolated from cells
- Can be reintroduced into a bacterial cell
- Small size(easy to manipulate and isolate)
- Circular (stable in the host cell).
- Several copies may be present(facilitates replication)
- Frequently have antibiotic resistance(detection easy).
Disadvantages:
- Cannot accept too large fragments (typical Sizes range from 0 – 10kb)
五、Other types of Vectors (besides plasmid)
Phage vectors
1. Introduction
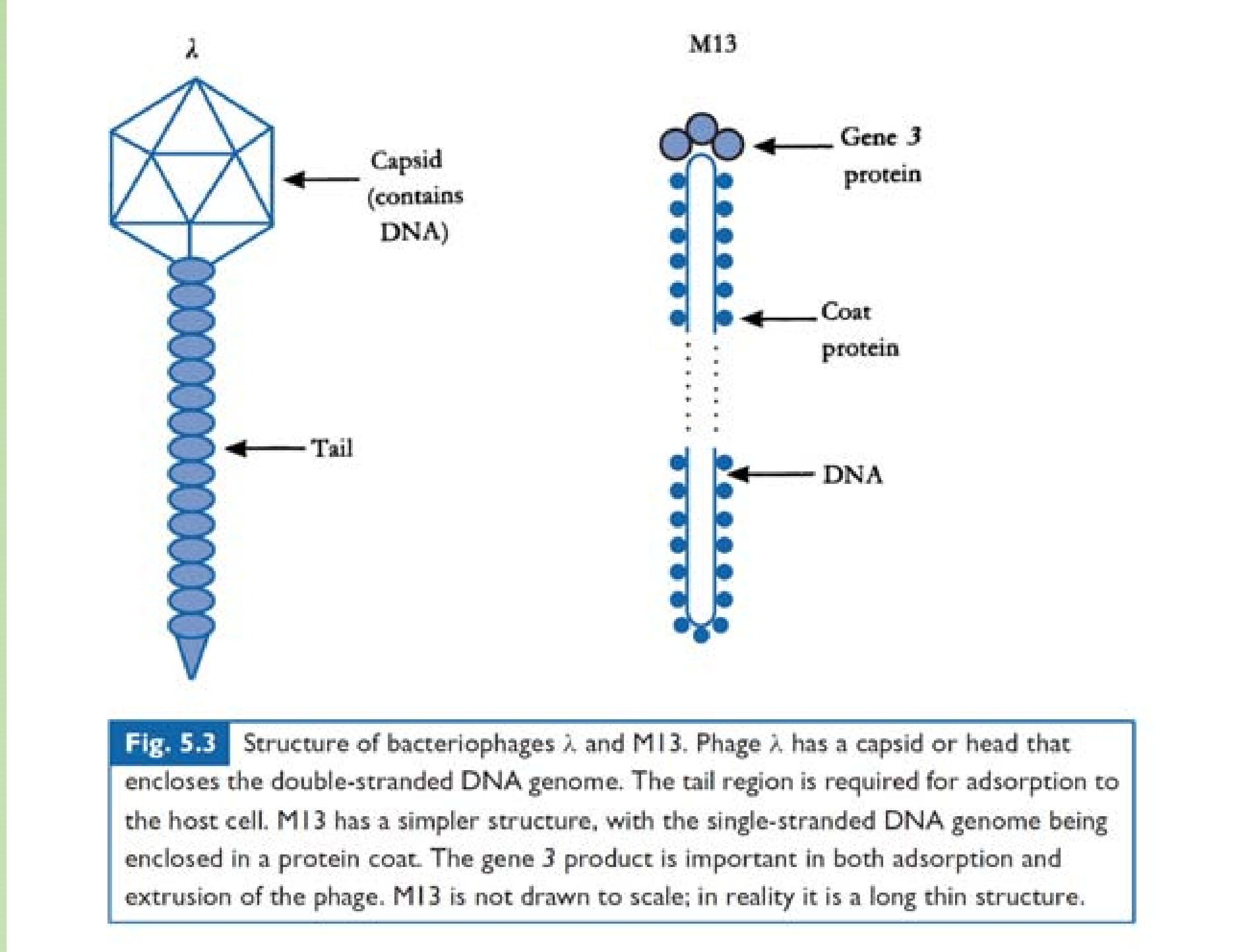
Phages are Bacteria eaters.
- tailless,
- head with tail
- filamentous.
Genome: ssDNA/dsDNA/RNA. dsDNA most often
Phages may be classified as either virulent or temperate, depending on their life cycles. Virulent under goes lytic growth only. While temperate phage under goes Lysogenic cycle mostly, but can also go to lytic growth some time.
lytic growth cycle or lysogenic cycle:
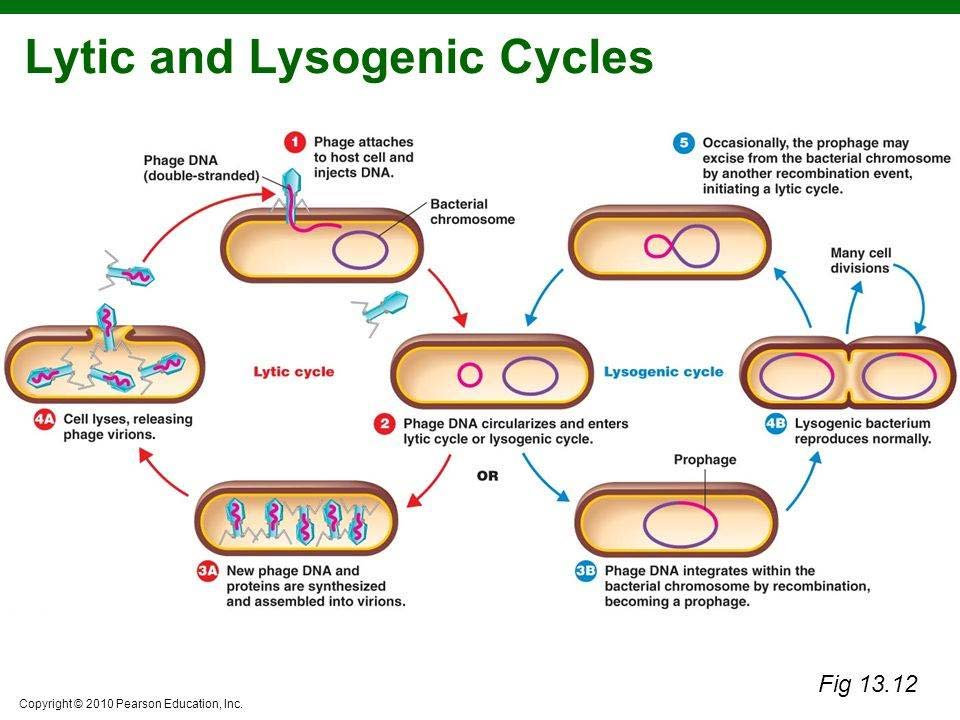
2. Why we might fancy Phage vectors ?
Derivatives of phage have been developed as cloning vectors since the earl of gene technology
The phage derivatives are considered to be the most suitable cloning vehicles for cloning genomic eukaryotic DNA because of the following advantages over the plasmids.
Easy to transform: thousands of phage plaques can be obtained in a single petri dish
Millions of independently cloned virus particle can be constituted to form a gene library
Selection by DNA-DNA hybridization is possible
3. Lambda Phage Vectors
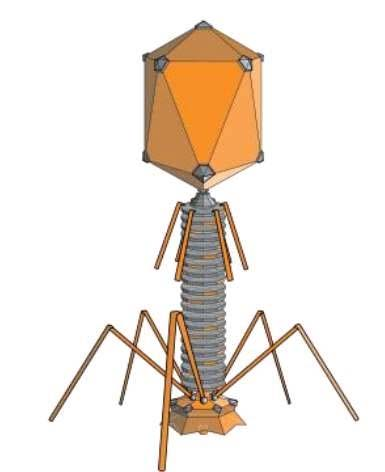
Lambda phage genome: a linear dsDNA of 48502 bp (~49kb) in length
The DNA isolated from virus particles is a double stranded linear molecule with short complementary single stranded projections of 12 nucleotides at its 5’ ends.
These cohesive termini, also referred to as cos sites, allow the DNA to be circularized after infection of the host cell.
The life cycle of lambda phage
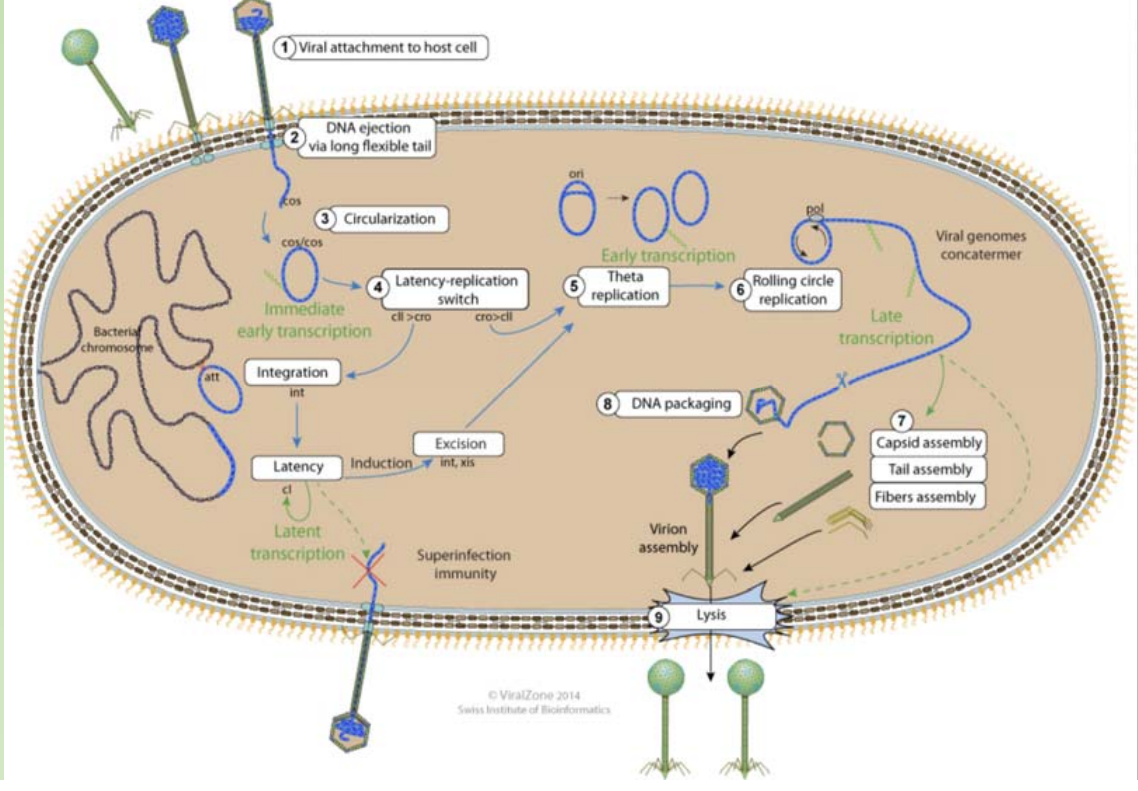
Adsorption: phage particle binding to receptors on the bacterial surface
Prophage: If the lysogenic cycle is initiated, the phage genome integrates into the host chromosome and is maintained as a prophage. If the lytic cycle is initiated, a complex sequence of transcriptional events essentially enables the phage to take over the host cell and produce multiple copies of the genome and the structural proteins. These components are then assembled or packaged into mature phage, which are released following lysis of the host cell.
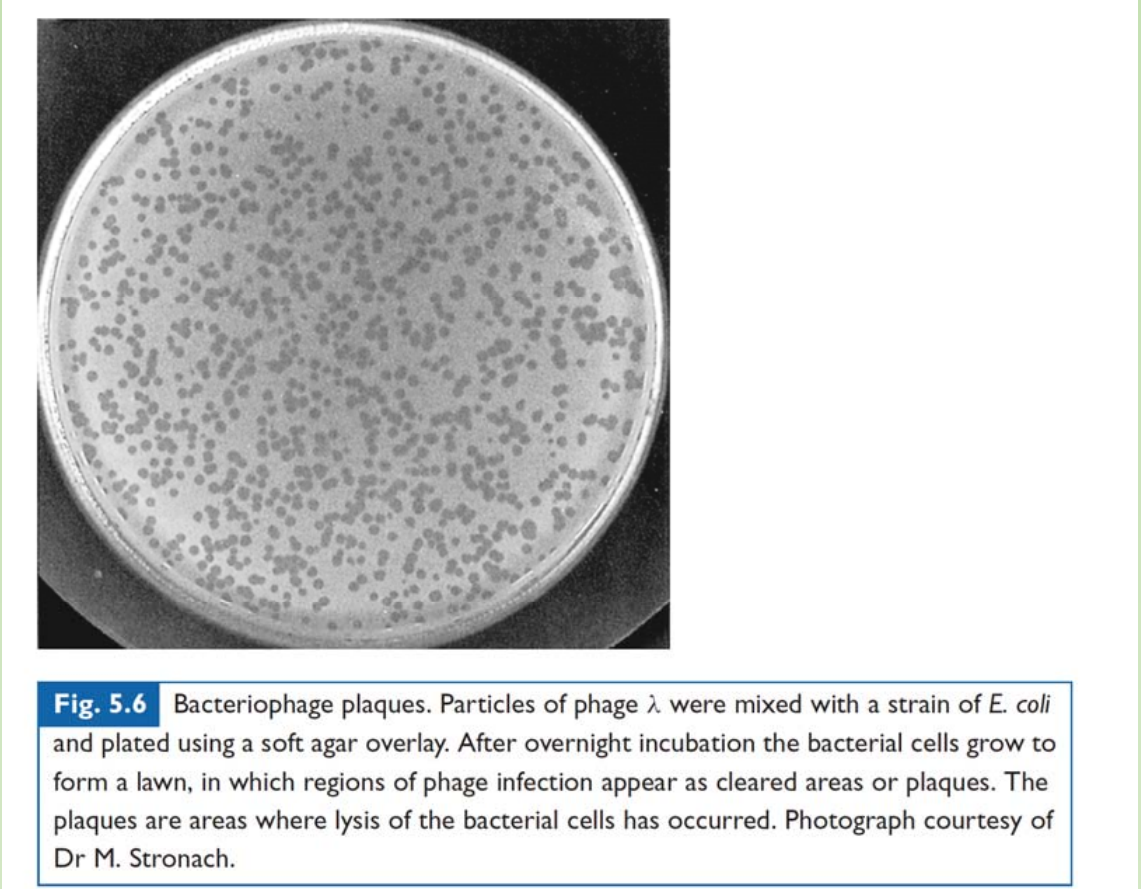
Plaque: Particles of phage λ were mixed with a strain of E. coli and plated using a soft agar overlay. After overnight incubation the bacterial cells grow to form a lawn, in which regions of phage infection appear as cleared areas or plaques. The plaques are areas where lysis of the bacterial cells has occurred.
GENERAL STRUCTURE OF BACTERIOPHAGE
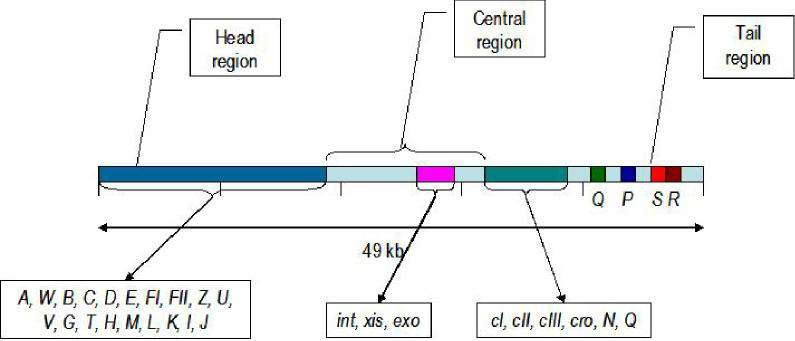
The genetic map of phage comprises approximately 40 genes which are organized in functional clusters.
Genes coding for head and tail are proteins (genes A-J) are on the left of the linear map
The central region contains genes, such as int, xis, exo etc., which are responsible for lysogenisation i.e the process leading to the integration of viral DNA and other recombination events. - Much of this central region is not essential for lytic growth.
Genes to the right of the central region comprise six regulatory genes, two genes (O and P) which are essential for DNA replication during lytic growth and two more genes (S and R) which are required for the lysis of the cellular membranes
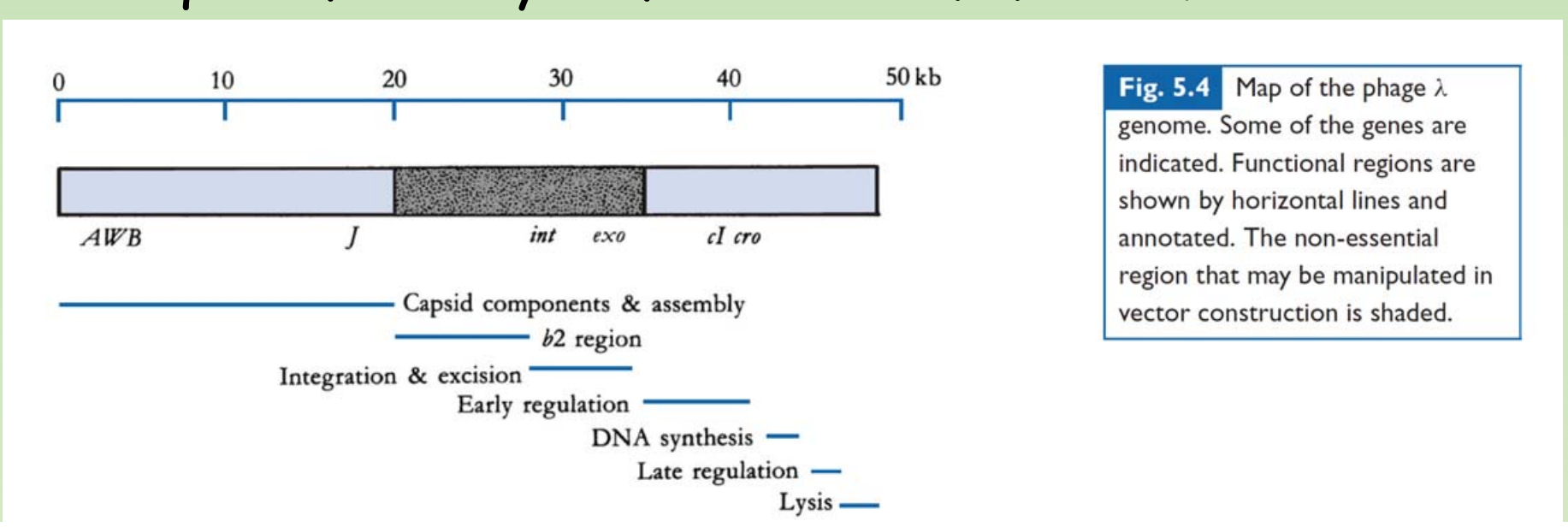
In the phage DNA, larger central region is not essential for phage growth and replication.
- This region of phage can be deleted or replaced without seriously impairing the phage growth cycle.
- Using this non-essential region of phage, several phage vector derivatives have been constructed for efficient gene cloning.
TYPES OF PHAGE VECTORS
Wild type phage DNA itself cannot be used as a vector since it contains too many restriction sites.
Further, these sites are often located within the essential regions for phage’s growth and development
From these wild phages, derivatives with single target sites and two target sites have been synthesized.
Phage vectors which contain single site for the insertion of foreign DNA have been designated as Insertional vectors.
Vectors with two cleavage sites, which allow foreign DNA to be substituted for the DNA sequences between those sites, are known as replacement vectors
1. INSERTIONAL VECTORS
A large segment of the non-essential region has been deleted, and the two arms ligated together.
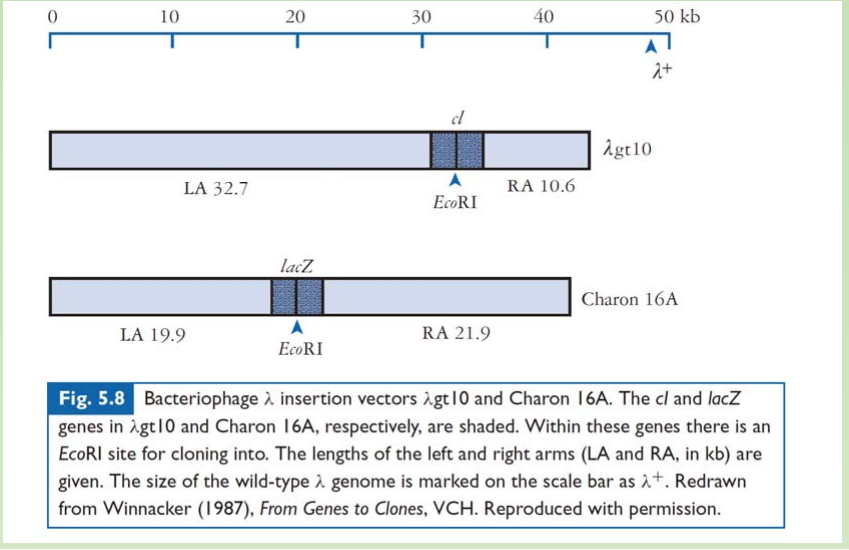
- A large segment of the non-essential region has been deleted, and the two arms ligated together
- An insertion vector possesses at least one unique restriction site into which new DNA can be inserted
- Capsid places a physical constraint on the amount of DNA that can be incorporated
- During phage assembly. viable phage particles can be produced from DNA that is between approximately 38 and 51 kb in length.
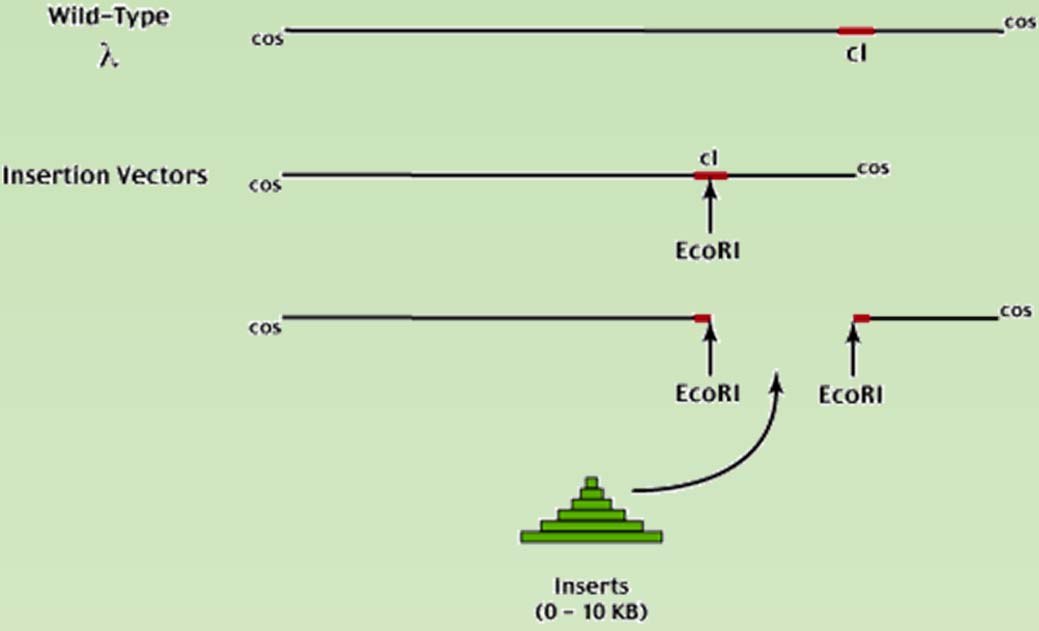
Selection system for the insertional lambda phage vector.
LacZ – blue/write selection is one way.
cl gene – product is necessary for the lysogenic growth of the phage.
- Insertion into the cl gene would leads to the phage go strictly through the lytic cycle
- WT phage would go to the lysogenic growth when infect E.coli that had an prophage already, the infection of the phage would leads to the slow growth but not lysis of cell, which would leads to cloudy but not transparent plaque.
- Recombinant phage that with cl gene disrupted would be forced to go for lytic cycle, which would leads to the transparent plaque.
Wild-type lambda phage do not efficiently infect E coli lysogenic for an unrelated bacteriophage P2.
- Thus, wild-type phage are Sensitive to P2 Interference (spi).
- Phage genes responsible for the spi phenotype are carried on the stuffer fragment of the insertion vector.
- Removal of the stuffer makes the recombinant phage insensitive to P2 interference
- Therefore, only the recombinant can growth in the presence of P2.s
2. REPLACEMENT VECTORS
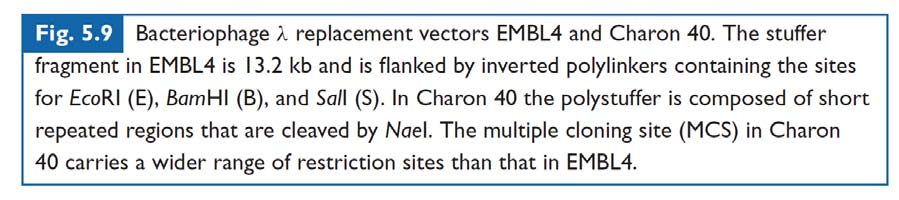
Replacement vector - foreign DNA replaces a piece of DNA (stuffer fragment) of the vector. Compared with insertional vector, longer DNA can be inserted.
- A central ‘stuffer’ fragment is removed and replaced with the insert DNA.
- DNA fragments between approximately 9 and 22 kb may be cloned in EMBL4;
- PolyStuffer: stuffer made by multiple repeats of short piece of DNA – to increase the cloning efficiency (as the NaeI will cut the stuffer into pieces).
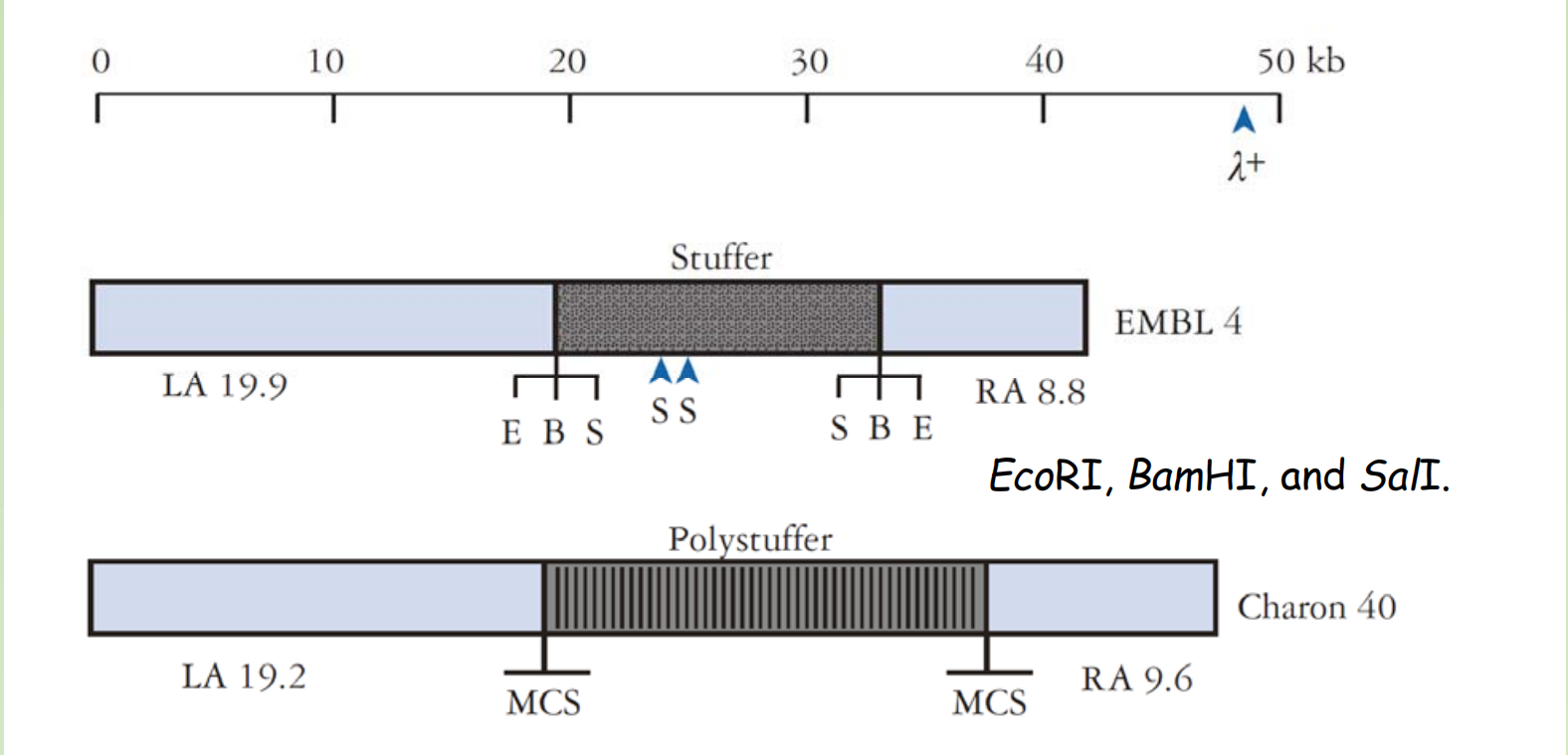
How do we transform phage vector into E. coli ?
Key benefit of phage vector: transduction was done through infecting the E.coli with in-vitro packed phage particles
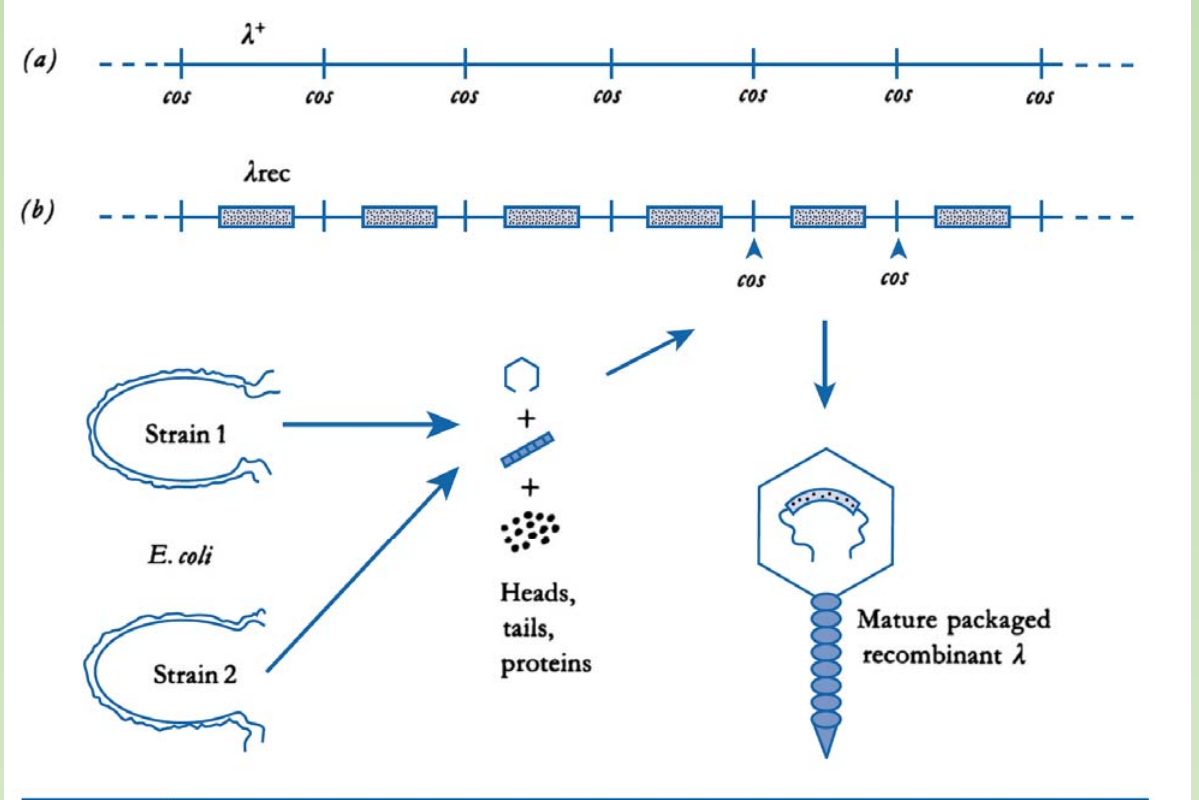
- There are strains (1&2) supplement proteins required for packaging of phage particles
- In this case, only the cos site is required for the packaging of DNA into the capside.
Cosmid
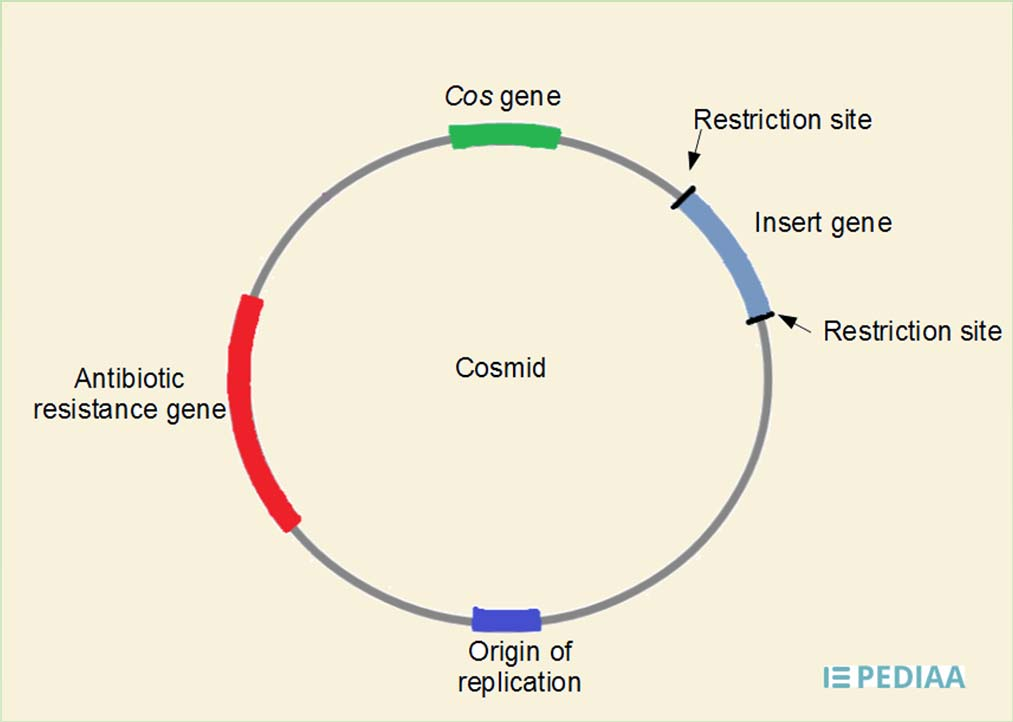
Cosmid is a hybrid of plasmid and lamda phage vectors
Plasmid with lamda phage packaging sequence(cos)
Cosmid are able to contain 37 to 52kb of DNA
Packaged into lamda phage particles and injected into host cells
Circularizes in cell and continues as a large plasmid
BACTERIAL ARTIFICIAL CHROMOSOMES (BAC)
BACs are based on bacterial mini-F plasmids, which are small pieces of episomal bacterial DNA that give the bacteria the ability to initiate conjugation with adjacent bacteria. They have a cloning limit of 75-300 kb
BAC is named after the YAC, but it is not actually maintained as chromosome.
Actually, the Human genome project mainly take advantage of BAC
YEAST ARTIFICIAL CHROMOSOMES (YAC)
1. Introduction
YACs are artificial chromosomes that replicate in yeast cells. They consist of Telomeres, which are ends of chromosomes involved in the replication and stability of linear DNA.
Origin of replication sequences necessary for the replication in yeast cells.
A yeast centromere, which is a specialized chromosomal region where spindle fibers
- attach during mitosis
A selectable marker for identification in yeast cells
- Ampicillin resistance gene for selective amplification
- Recognition sites for restriction enzymes
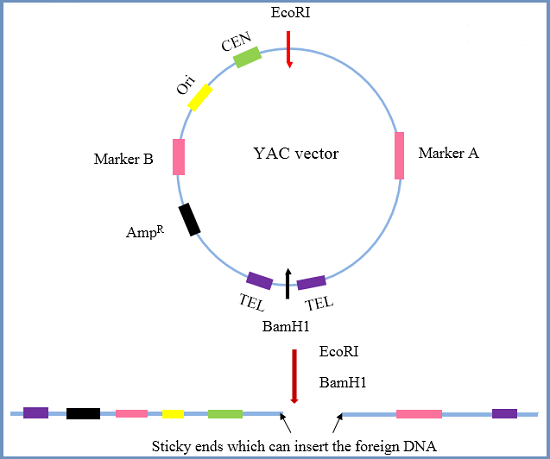
2. THE PROCEDURE FOR MAKING YAC VECTORS
- The target DNA is partially digested by a restriction endonuclease, and the YAC
vector is cleaved by restriction enzymes. - The cleaved vector segments are ligated with a digested DNA fragment to form
an artificial chromosome. - Yeast cells are transformed to make a large number of copies.
They are the largest of the cloning vectors, with a cloning limit of 100-1000 kb, however they have very low efficiency
SHUTTLE VECTORS
Definition: They are the vectors that can shuttle between more than one host, for example, one is E. coli and the other is yeast.
Structure and function: Most of the vectors for use in eukaryotic cells are constructed as shuttle vectors
In E. coli:
- • This means that they can survive and have the genes (ori and ampr ) required for replication and selection in E. coli.
In the desired eukaryotic cells:
- They can also survive in the desired host cells, and let the target insert sequences take effects.
E.coli EXPRESSION VECTORS
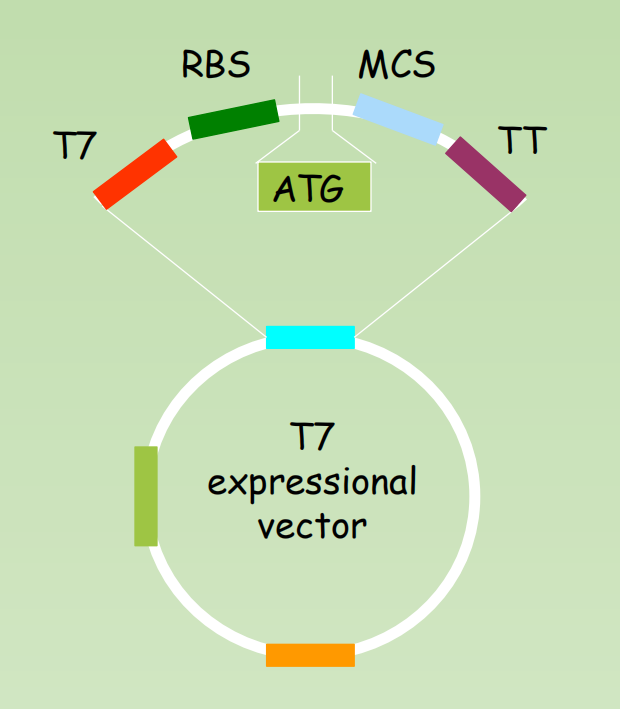
Definition of expression vectors: Cloned genes – expression vector – hostfusion protein expression
Structure:
- Promoter: a strong promoter (T7)
- RBS: Ribosome binding site
- ATG: translation initiation condon
- MCS: Multiple cloning sites
- TT: transcription terminator
- ampr, Ori,
- Tag for affinity purification: His-tag, GST, MBP etc