L11 Biological Catalysts
一、Overview
The Role of Enzymes
1. Catalysis
In general terms, a catalyst is a substance that increases the rate, or velocity, of a chemical reaction without itself being changed in the overall process.
Catalysis is essential to make most critical biochemical reactions proceed at useful rates under physiological conditions
Example:
Hydrogen peroxide
$$
2\ H_{2}O_{2} \rarr 2\ H_{2}O+O_{2}
$$Bacterium reproduce in 20 minutes, without enzyme, it would be much slower
A human nerve cell must respond instantly to a stimulus, this process is greatly depends on catalysis
Features
Although a true catalyst participates in the reaction process, it is unchanged by the process.
Catalysts change rates of processes but do not affect the position of equilibrium for a reaction. A thermodynamically favorable process is not made more favorable, nor is an unfavorable process made favorable, by the presence of a catalyst. The equilibrium state is just approached more quickly.
This means that the catalyst can only change the kenetic but not the thermodynamics
(1) Specificity 特异性
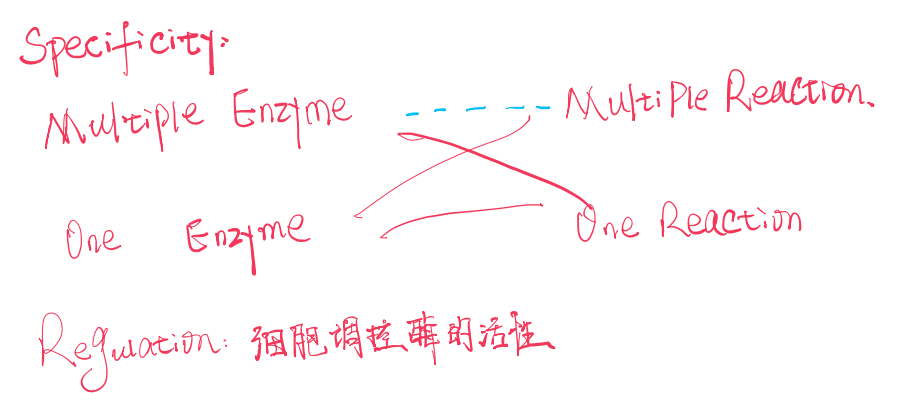
Multiple enzyme can catalyze one reaction;
One enzyme can catalyze multiple reactions or one specific reaction.
(2) Regulation
细胞控制酶分子的活性in different environment or states
2. Enzymes – Catalysts
Enzymes are biological catalysts, most of which are proteins.
- For example, the protein Trypsin catalyzes hydrolysis of peptide bonds in proteins and polypeptides.
The substance that is acted on by an enzyme is called the substrate(底物) of that enzyme.
Function
(1) Alter The Reaction Pathway
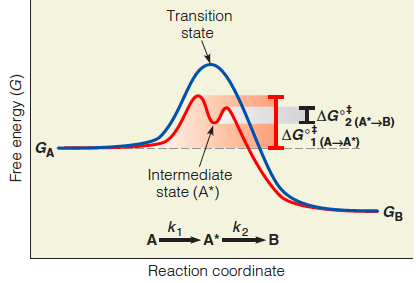
An enzyme may alter the reaction pathway to one that includes one or more intermediate states(中间态) that resemble (类似于) the transition state(过渡态) but have a lower free energy.
In the case of a single intermediate, the activation energies for formation of the intermediate state and for conversion of the intermediate to product are lower than the activation energy for the uncatalyzed reaction.
Covalent bond involves in the intermediate state.
(2) stabilizing an intermediate states
(3) More about the function
Enzymes do more than just simply distort or position their substrates
binds the substrate or substrates
lowers the energy of the transition state
directly promotes the catalytic event
Specific amino acid side chains
Specific amino acid side chains poised (处于(危险的)不稳定的状态之中) in exactly the right places to aid in the catalytic process itself.
In many cases, these side chains are acidic or basic groups that can promote the addition or removal of protons.
In other instances, the enzyme holds a metal ion in exactly the right position to participate in catalysis.
Active Site
In an enzyme, a site is complementary in shape, charge, and polarity to the transition state for the reaction, and to a lesser extent, its substrate.
- Which is also called Substrate-binding site
Enzyme and its substrate has complementarity(互补性)
- This complementary bind is caused by weak interactions
- This is the basis for the specificity of enzyme-catalyzed reactions.
- Transition state is more preferred than substrate of enzyme
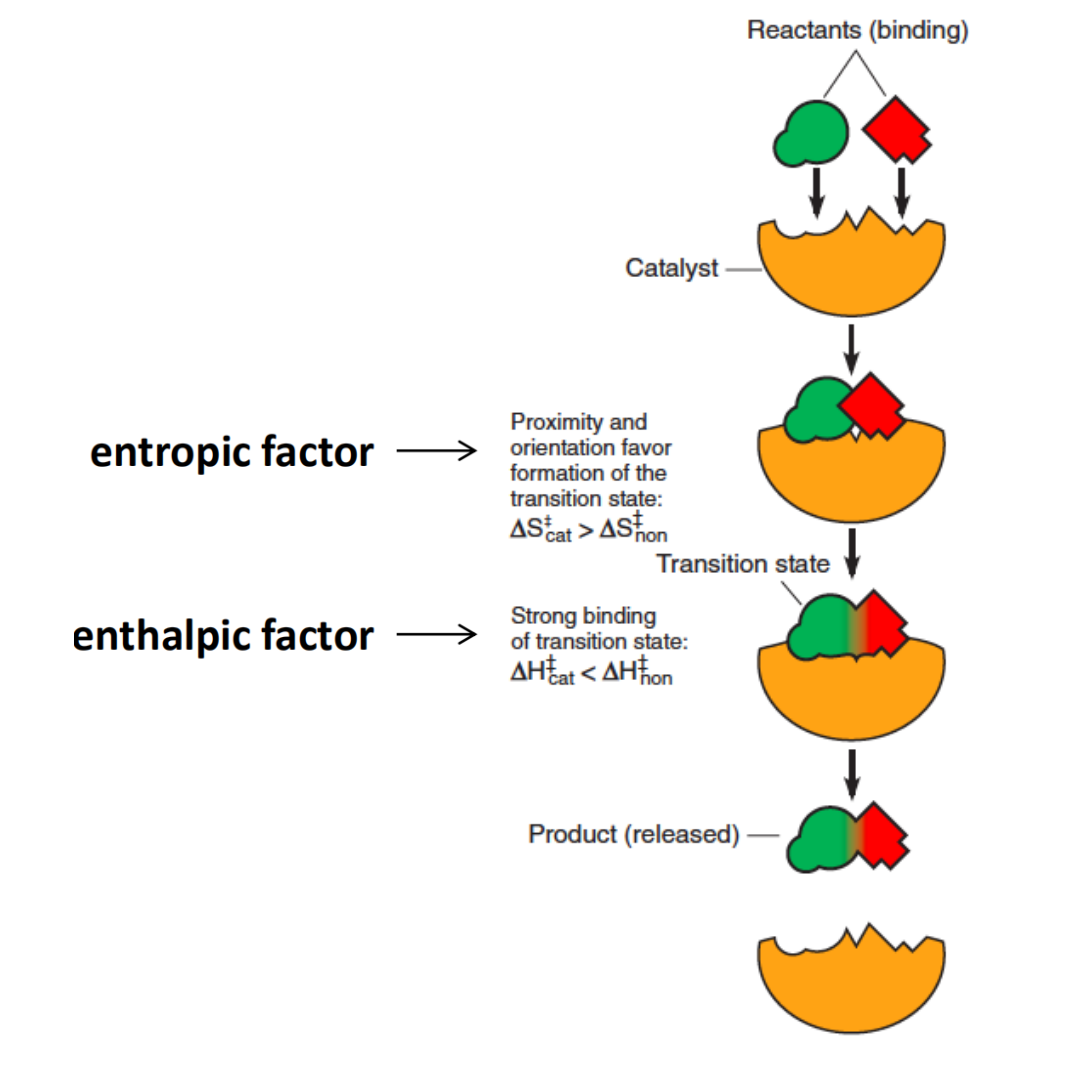
3. Entropic and enthalpic factors in Catalysts
Two reactants are bound to sites on the catalyst which ensures their correct mutual orientation and proximity, and binds them most strongly when they are in the transition state conformation.
Chemical reaction rates
The order of reaction:
$$
v = k[A]^{x}[B]^{y} \ \text{k = rate constant} \ \text{For reaction} \ A \rarr B
$$
The larger value of $k_{1}$, the more rapid the rate, and vice versa.
For first order reaction:
$$
v = -\frac{d[A]}{dt}\ \because \int^{[A]{t}}{[A]{0}}\frac{d[A]}{[A]} = -k \int^{t}{0}{dt} \ \therefore [A]{t} = [A]{0}e^{-kt}
$$
Actually, A → B is a reversible reaction, so it is:
$$
A \xtofrom[k_{-1}]{k_{1}} B
$$
The equilibrium constant:
$$
v = -\frac{d[A]}{dt} = k_{1}[A]^{n}-k_{-1}[B]^{m} = \frac{d[B]}{dt}
\
\therefore k_{1}[A] = k_{-1}[B]
$$
$$
K = \frac{[B]}{[A]} = \frac{k_{1}}{k_{-1}}
$$
1. Half Life $t_{\text{1/2}}$
Half life is the time needed for [A] to decrease by one half.
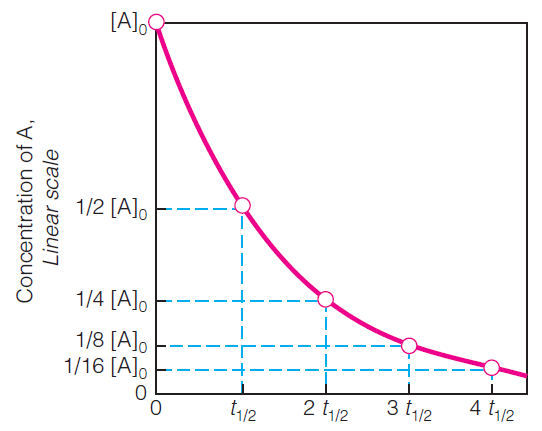
Smaller half life, the quicker the reaction is.
二、Catalysts and Chemical Reaction
Transition States and Reaction Rates
Barriers to chemical reactions occur because a reactant molecule must pass through a high-energy transition state to form products. This free energy barrier is called the activation energy(活化能).
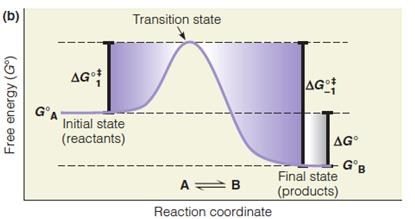
1. Dagger $\Dagger $
其中 $\Dagger$ (Dagger)指代的是Transition State or Activated State.
2. Transition State
The transition state is thought of as a stage through which the reacting molecule or molecules must pass, often one in which a molecule is strained or distorted or has a particular electronic structure, or in which molecules collide productively.
Increase The Reaction Rate
1. Standard Free Energy of Activation
$$
\text{The standard free energy of activation: } \ \Delta G^{\circ \Dagger}
$$
This represents the additional free energy (above the average free energy of reactant molecules) that molecules must have to attain the transition state.
2. Catalyze Strategies
提升反应速率有两种策略:
- raising the temperature of the reaction
- lowering the free energy of the transition state.
Effect of increasing temperature or lowering $\Delta G^{o \Dagger} $ on the rates of reactions:
The rates of reactions are proportional (成比例的) to the number of molecules with sufficient energy to overcome the activation barrier $\Delta G^{o \Dagger} $
$$
\frac{[A^{\Dagger}]}{A} = e^{(\frac{-\Delta G^{0 \Dagger}}{RT})}
$$
$$
[A^{\Dagger}] \text{represent the fraction of molecules with sufficient energy to attain the transition state} \
[A] \text{is the fraction of A that remains in the reactant state}\
\text{ T is temperature and R is the gas constant}
$$
We know that in any sample or solution, not all molecules have the same energy at any instant
- 如果降低energy barrier, 那么对比原有的状态,降低了反应条件,因此满足活化能条件的分子就更多。
- A larger fraction of molecules will possess sufficient energy to attain the transition state, and the rate of reaction will increase
- 如果升高温度,那么对比原有状态,提升了一部分分子具有的能量,使满足反应条件的分子数目变多。
- As temperature increases the average kinetic energy of the molecules in a sample increases
So one way to change the reaction rate is change $\Delta G^{o \Dagger} $ and the temperature (T)
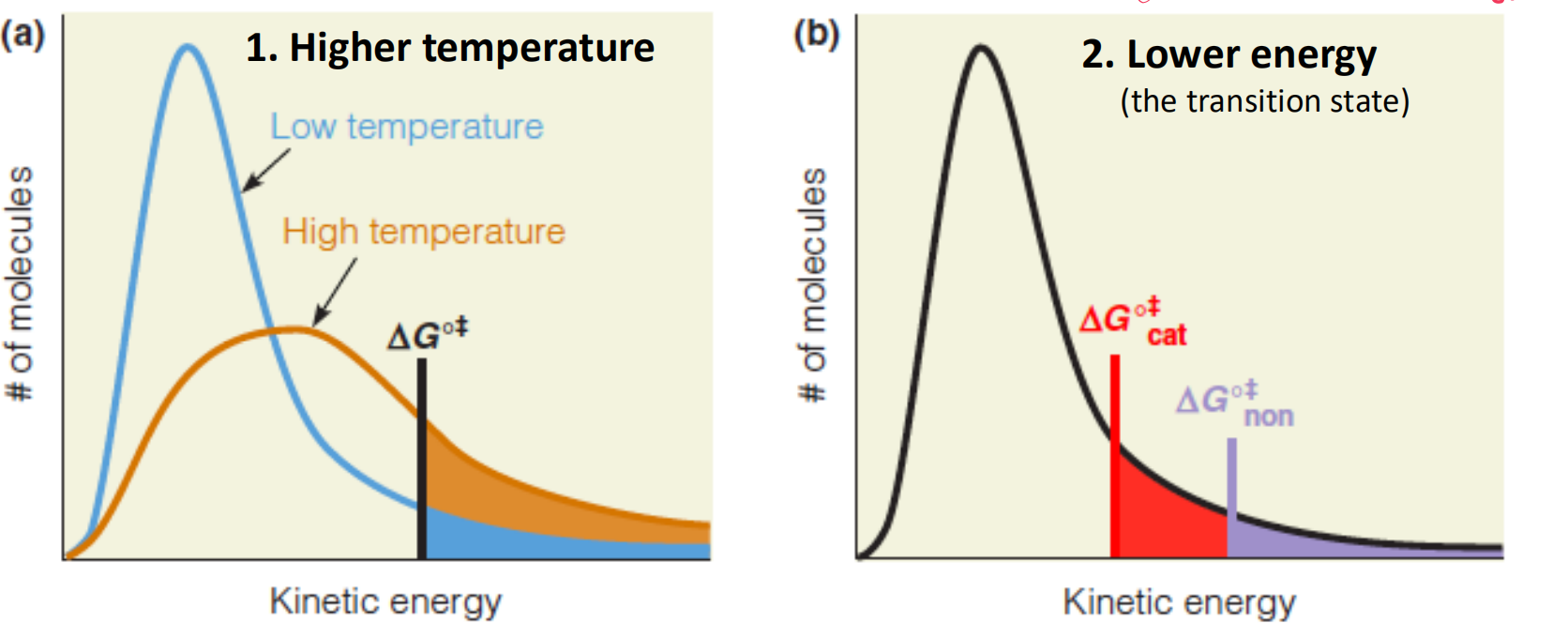
Transition state theory applied to catalysis
1. Rate Constant k
In its simplest formulation, transition state theory assumes that a reactant molecule that attains the transition state rapidly decomposes to a lower energy state such as the product state, or to an intermediate state
The rate enhancement for an enzyme-catalyzed reaction is the ratio of the rate constants for the catalyzed ($k_{cat}$) and the noncatalyzed ($k_{non}$) reactions.
$$
k = \gamma (\frac{k_{B} T}{h})e^{\frac{- \Delta G^{\circ \Dagger}}{RT}}
$$
where $k_{B}$ is the Boltzmann constant, h is Planck’s constant, T is temperature, R is the gas constant, and $\gamma$ is called the “transmission coefficient.”
The term $(k_{B}T)/h$ is a factor that accounts for the vibrational frequency of bonds in the transition state. At 310K (37℃), $(k_{B}T)/h$ has a value of ~$6.4 \times 10^{12} s^{-1}$
The value of $ \gamma$ for various enzyme reactions has been calculated from quantum mechanical principles and is often very close to 1 but may be as large as ~$10^3$ for some hydrogen transfer reactions where quantum tunneling is significant.
2. Rate Enhancement (Rate Acceleration)
The rate enhancement is the ratio of the rate constants for the catalyzed ($k_{cat} $) and the noncatalyzed ($k_{non}$) reactions for a given set of conditions (e.g., temperature, pH*,* etc*.*).
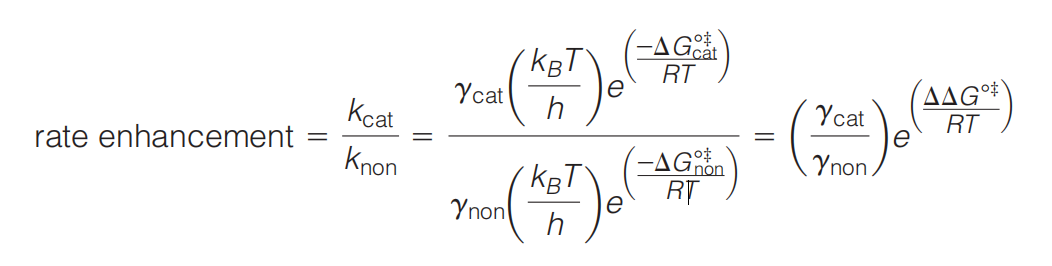
The rate enhancement indicates how much faster the reaction occurs in the presence of the enzyme.
- The rate enhancement is related to $\Delta \Delta G^{o \Dagger} $
The $\Delta \Delta G^{o \Dagger} $ , is the difference in activation energies between the noncatalyzed and catalyzed reactions
$$
\Delta \Delta G^{o \Dagger} = (\Delta G^{o \Dagger}{non} - \Delta G^{o \Dagger}{cat} )
$$
$\Delta \Delta G^{o \Dagger} $ indicates by how many kJ/mol the transition state is stabilized in the catalyzed reaction.
A catalyst lowers the activation energy barrier in both directions to the same extent. Thus, the rate accelerations for the forward and reverse reactions are identical. $\Delta \Delta G^{o \Dagger} $ unchanged
Effect of a catalyst on activation energy barrier
1. Enzyme choose lower the energy barrier
- The catalyst lowers the standard free energy of activation, $\Delta G^{o \Dagger} $
- 在生物体内不通过升高温度来提升反应速率的原因是:
- most organisms are sensitive to even small increases in temperature
- exothermic reactions become less favorable at elevated temperature.
- 在生物体内不通过升高温度来提升反应速率的原因是:
- The rate enhancement is related to $\Delta \Delta G^{o \Dagger} $ (Will explain it later)
- The reaction equilibrium is not perturbed by the presence of the catalyst
- $\Delta G^{o}_{A \rarr B} $ for both the catalyzed and noncatalyzed reactions are the same.
- $\Delta G^{o \Dagger}_{cat} $for both reaction directions are decreased. 因此不改变化学平衡
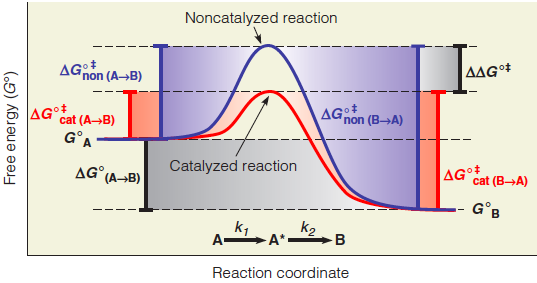
2. Strategies for Catalyze to lower the Barrier
已知:
$$
\Delta G^{\circ \Dagger} = \Delta H^{\circ \Dagger} - T \Delta S^{\circ \Dagger}
$$
所以有两种方式可以减少$\Delta G^{\circ \Dagger}$:
- Reduce $\Delta H^{\circ \Dagger}$ ($\Delta H^{\circ \Dagger}{cat} < \Delta H^{\circ \Dagger}{non}$)
- Increase $\Delta S^{\circ \Dagger}$ ($\Delta S^{\circ \Dagger}{cat} > \Delta S^{\circ \Dagger}{non}$)
Reduce $\Delta H^{\circ \Dagger}$
$\Delta H^{\circ \Dagger}$ can be lowered by increasing the number of bonding interactions between the catalyst and the transition state.
This appears to be the dominant effect in most enzyme-catalyzed reactions.
Increase $\Delta S^{\circ \Dagger}$
The $\Delta S^{\circ \Dagger}$ term reflects the fact that a particular orientation between reactants or parts of a molecule may be necessary to achieve the transition state.
A catalyst that can bind two reacting molecules in proper mutual orientation will increase their reactivity by making the entropy of activation less negative
(1) Strain 应变 and Conformational Change
In a first-order reaction, which involves something happening within a single molecule, parts of the molecule must often be re-oriented properly to allow the transition state to be reached. Such re-orientation is sometimes referred to as strain.
In the case of enzymes, which are structurally dynamic catalysts, it is not always clear whether conformational strain between the enzyme and substrate induces a larger change in the conformation of the enzyme or in the substrate.
The thermodynamic cost of making $\Delta S^{\circ \Dagger}$ more positive is paid by favorable enthalpic interactions between the catalyst and substrate.
Intermediate State
In some cases the catalyst can reduce the activation energy requirement by altering the reaction pathway and stabilizing an intermediate state that resembles the transition state but is of lower energy
(1) Difference Between Intermediate and Transition State
We distinguish such an intermediate state from a transition state by the fact that the intermediate state corresponds to a local free energy minimum, and the transition state to a free energy maximum.
三、How Enzymes Acts As Enzymes
Two models for enzyme–substrate interaction
In this example, the enzyme catalyzes a cleavage reaction.
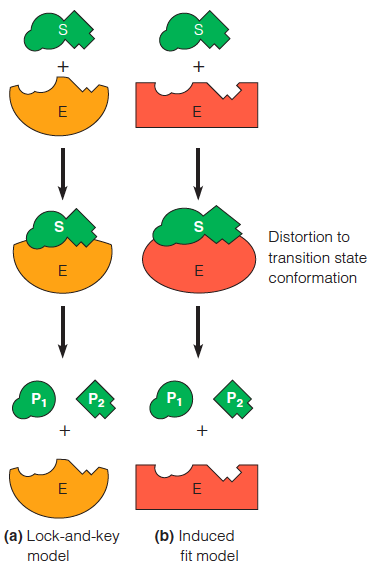
1. The lock-and-key model
In this early model, the active site of the enzyme fits the substrate as a lock does a key
2. The induced fit model
In this elaboration of the lock-and-key model, both enzyme and substrate are distorted on binding. The substrate is forced into a conformation approximating the transition state; the enzyme keeps the substrate under strain.
- Involves conformational change
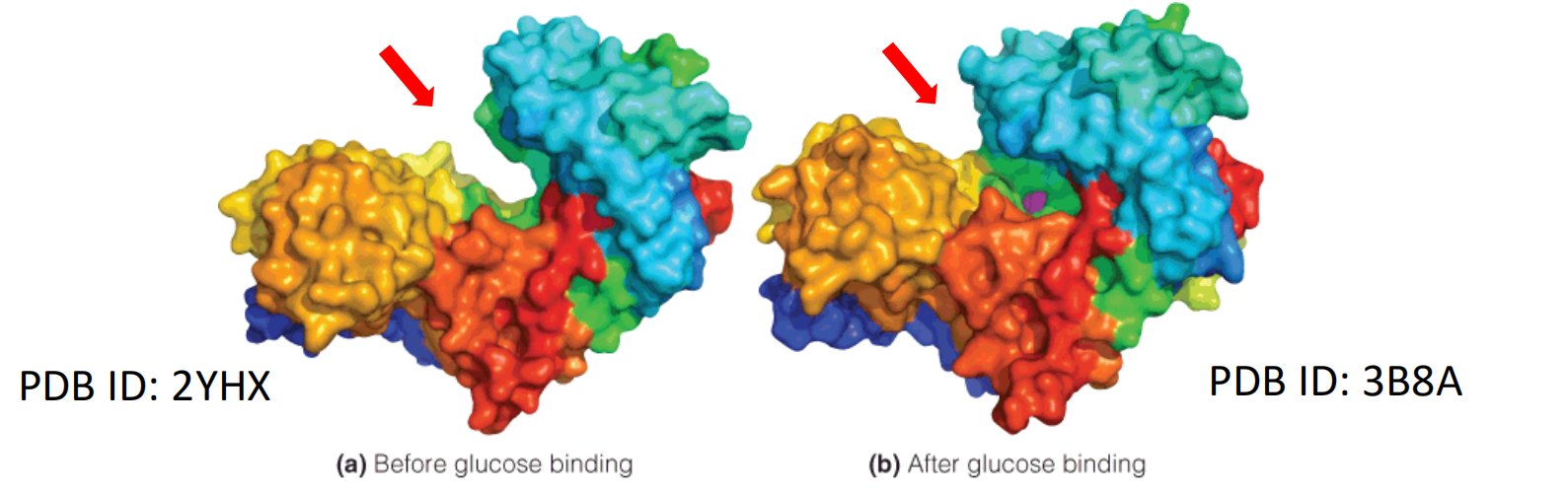
Example: The induced conformational change in hexokinase
The binding of glucose to hexokinase induces a significant conformational change in the enzyme.
The enzyme is a single polypeptide chain, with two major domains.
A simple enzyme-catalyzed reaction
1. The Relationship of $k$
For an enzyme (E) that catalyzes the conversion of a single substrate (S) into a single product (P), ES is the enzyme–substrate complex, and EP is the enzyme bound to the product.
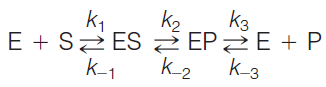
The expression for the reaction includes three steps:
The first step, binding of substrate, is reversible (i.e., $k_{1} \text{ and }k_{-1} >> k_{2}$).
The second step, conversion of ES to EP, lies far to the right (i.e., $k_{2} >> k_{-2}$ ).
The third step, release of product, is rapid compared to the catalytic step p (i.e., $k_{3} >> k_{2}$)
The three major assumptions above later on will be used to develop a rate expression (Michaelis-Menten equation).
2. Reaction coordinate diagram
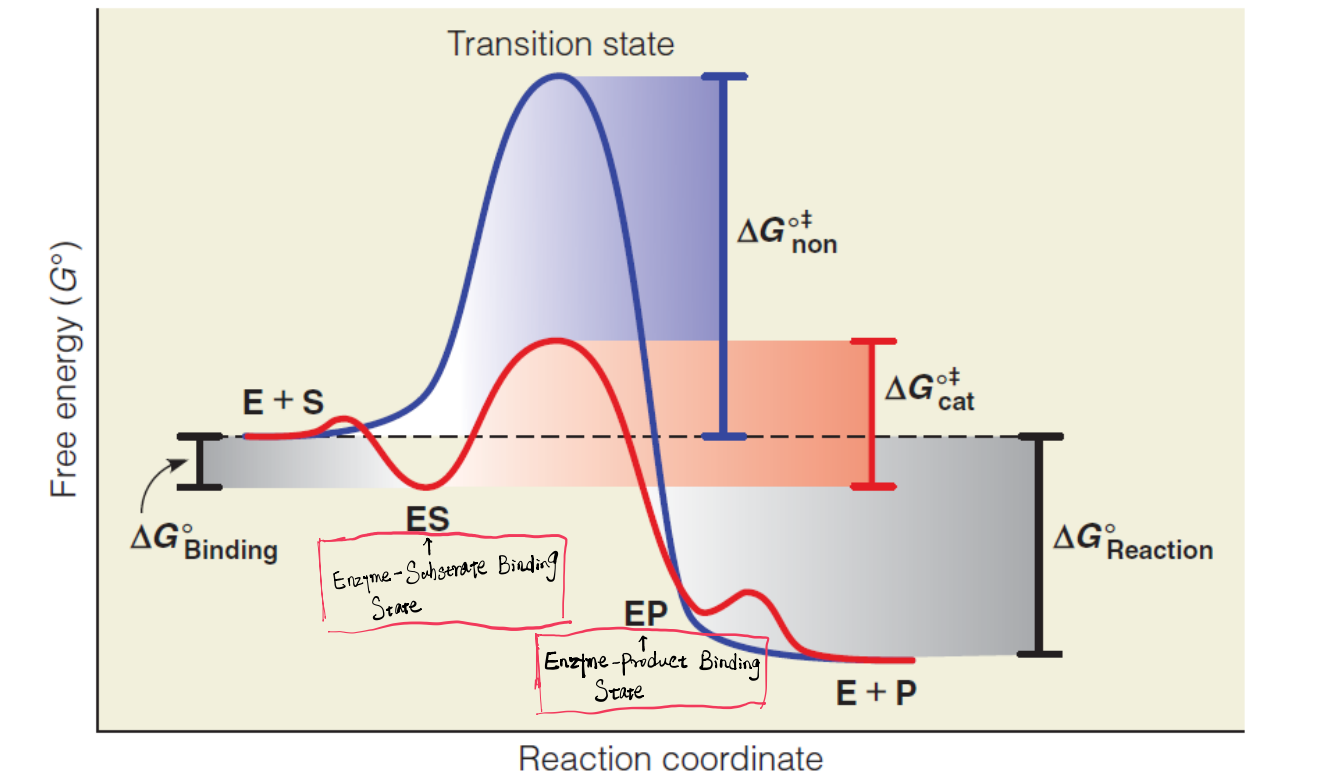
Formation of the ES complex tends to be thermodynamically favorable
- $E_{ES}<E_{E+S}$
Binding to product should be less favorable than its release from the active site (Release is favorable)
- $E_{EP}>E_{E+{P}}$
The active site binds the transition state more favorably than it binds the substrate
- $\Delta G^{o \Dagger}{cat} < \Delta G^{o \Dagger}{non}$
Energy barrier is small for both E+S → ES and EP → E+P
The Mechanisms of Enzyme
Binding to the transition state: electrostatic catalysis
- Via noncovalent bonding interactions (H-bonds, charge-charge interactions, etc.)
Distortion of substrate and/or active site to reduce activation energy: induced fit
Binding of substrates to optimize proximity and orientation: making $\Delta S^{o \Dagger}$ more favorable
Altering the reaction pathway to include intermediate states: covalent catalysis
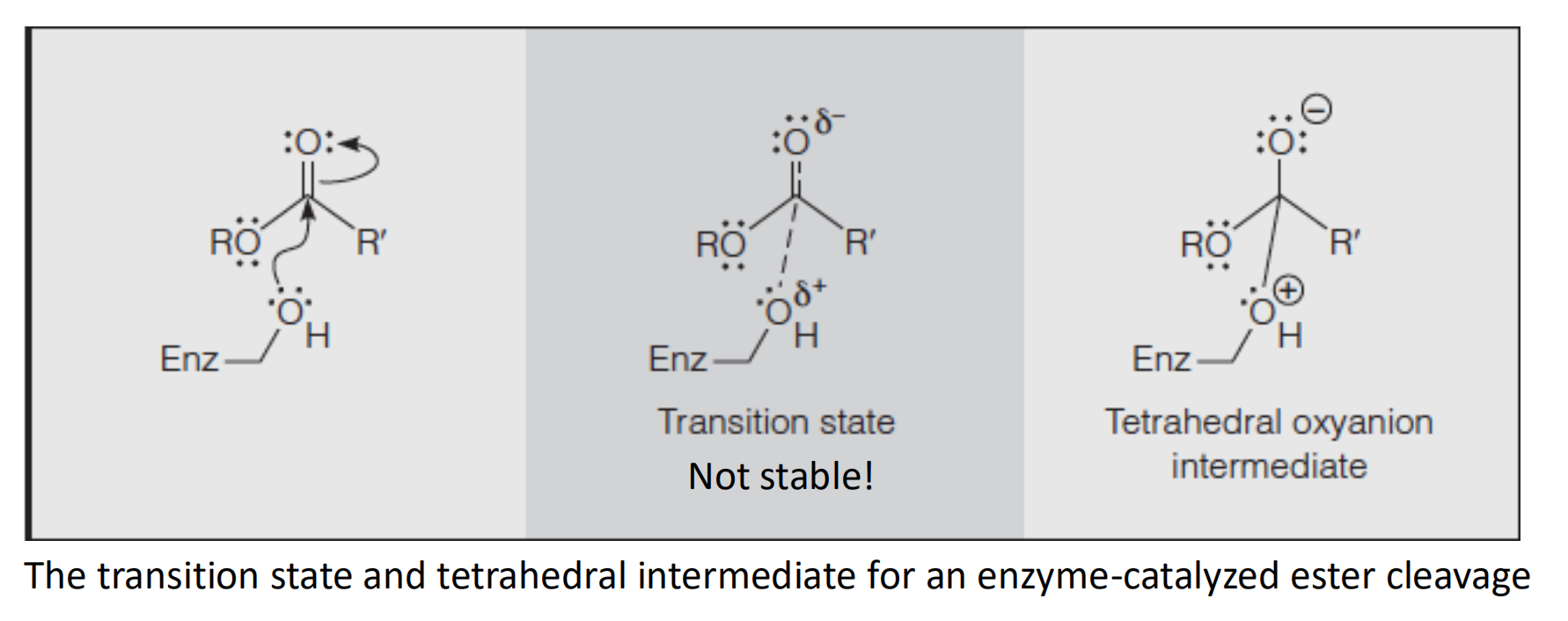
- General acid/base catalysis
- Metal ion catalysis
1. General acid/base catalysis
General acid/base catalysis (GABC) is important in reactions involving proton transfer.
General acid/base
- Proton donor/acceptor
- Donate/remove an proton to an atom that develops negative/positive charge in the transition state
- Polar residues
The transition state might be stabilized by electrostatic interactions with active site amino acids and/or metal ions, or it might be stabilized by general acids (GAC) or bases (GBC).
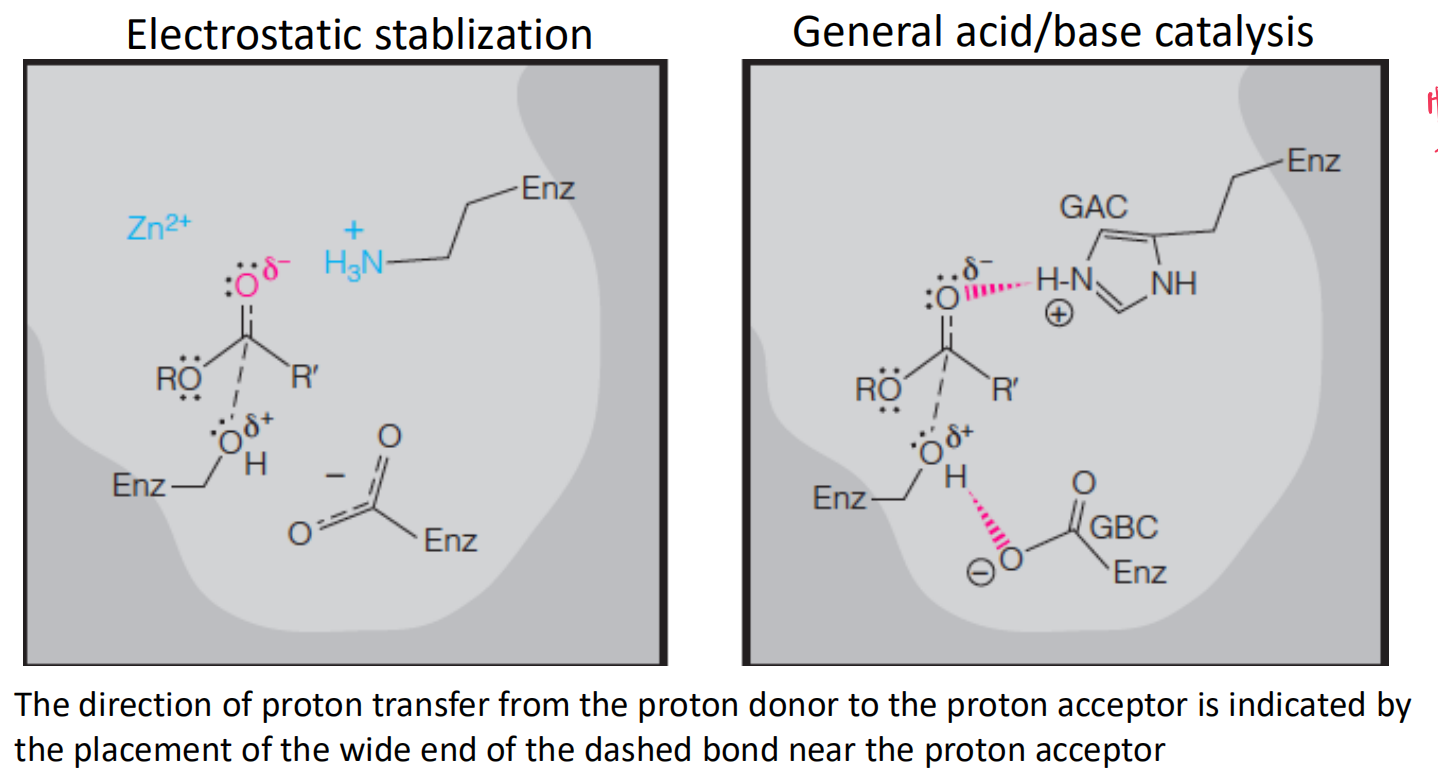
Electrostatic Stabilization与GABC的区别在于有没有proton的accept与donate
根本原因在于Side Chain的性质 – $pK_{a}$
右图中发生proton donate/accept的是His,由于His的$pK_{a}$较小,在pH=6的情况下就可以发生deprotonalized
Case 1: Lysozyme 溶菌酶
Lysosome is an enzyme that cleaves the peptidoglycan layer of gram-positive bacteria
(1) The active site cleft of lysozyme:
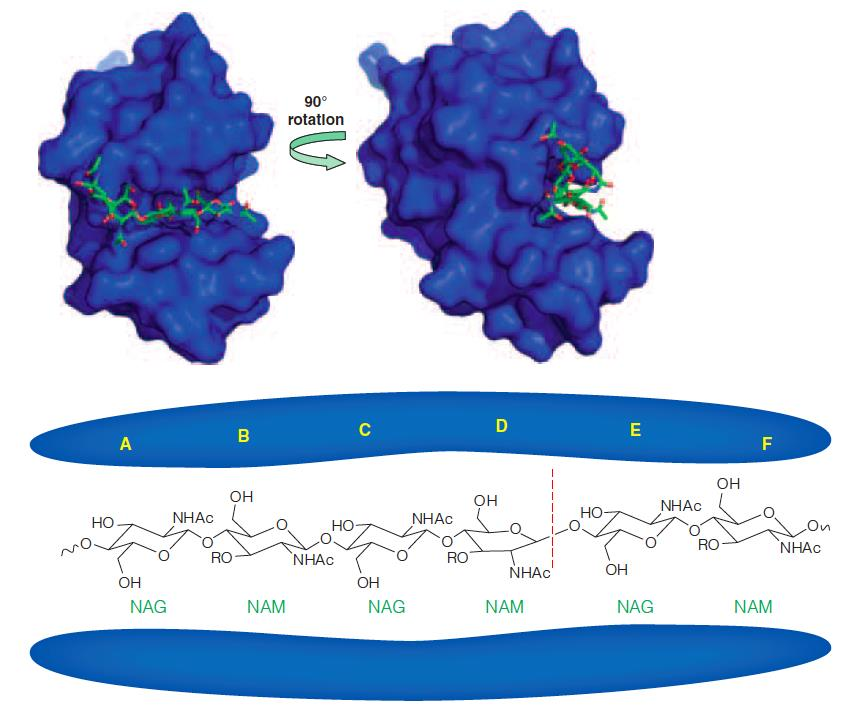
Top: The solvent-accessible surface of hen lysozyme is shown in blue.
The trisaccharide(三糖) NAM-NAG-NAM is shown in stick representation bound to the active site.
Bottom: A schematic drawing of $(NAG-NAM)_{3}$bound to the A–F subsites in lysozyme.
The active site of lysozyme is located in a deep cleft (Figure 11.13) where six glycosyl residues bind in subsites labeled A–F.
The glycosidic bond cleavage occurs between residues D and E, with retention of configuration at the anomeric carbon (i.e., C1 of the pyranose in the D subsite).
(2) The mechanism of action of lysozyme:
There are some given mechanisms (hypothesis)
Phillips mechanism
The Phillips mechanism is illustrated by the black reaction arrows along the left side of the diagram.
Phillips mechanism 在下图中以黑色箭头表示
Phillips proposed a stereochemical mechanism in which glutamic acid 35 (E35) acts as a general acid and aspartate 52 (D52) acts as an electrostatic catalyst during the generation of an oxocarbenium ion in the transition state
In the first step, E35 acts as a general acid to promote cleavage of the glycosidic bond and concomitant formation of the oxocarbenium ion (which is stabilized electrostatically by D52).
In the second step, E35 acts as a general base, deprotonating a water molecule, which then attacks C1 of the substrate.
Phillips also proposed that the residue in the D subsite is distorted into a half-chair conformation upon binding in the active site (see Figure 11.2). This conformation approximates that of the oxocarbenium ion and is an example of the substrate distortion (“strain”) discussed above.
The Phillips mechanism proposes that D52 stabilizes the oxocarbenium ion via electrostatic stabilization and also shields one side of the ion from attack by water, thus explaining the retention of configuration at the anomeric carbon (C1).
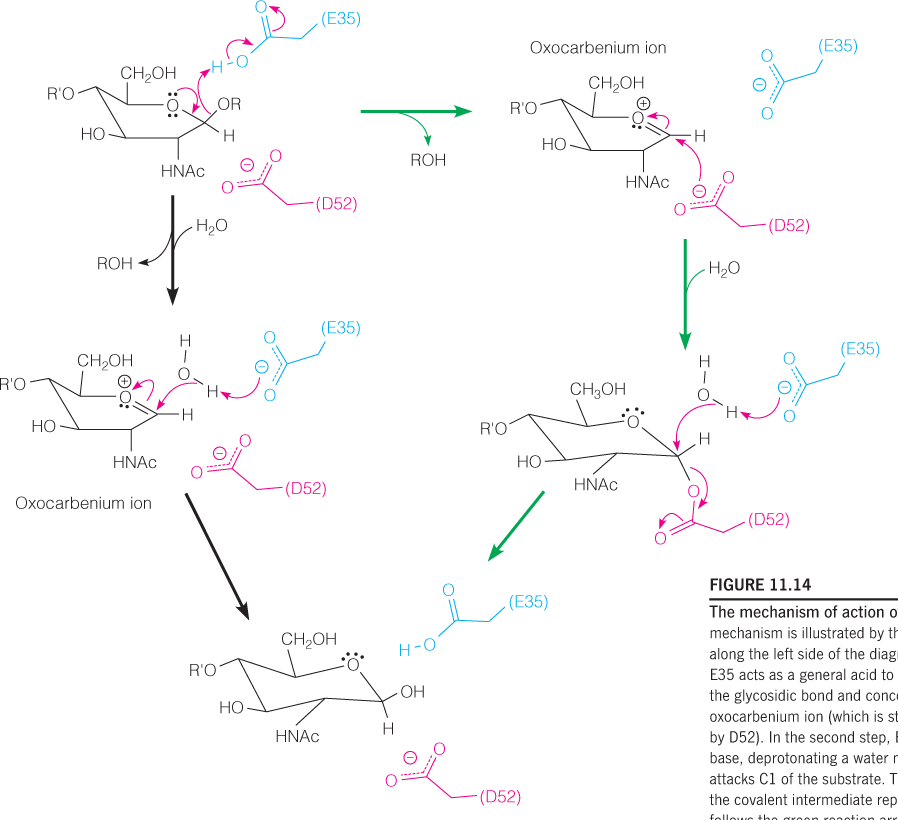
Steve Withers Mechanism
The pathway that includes the covalent intermediate reported by Steve Withers follows the green reaction arrows along the right side of the diagram.
In 2001, Steve Withers’ laboratory reported a crystal structure of an enzyme-glycosyl intermediate that showed the D52 side chain covalently bound to C1
In this case, the second step involves covalent bond formation between C1 of the substrate and D52.
Attack of the water displaces D52 in the subsequent step.
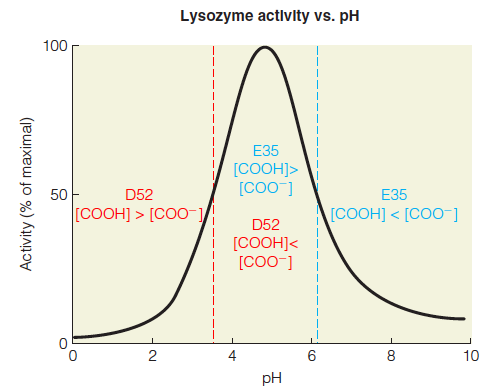
(3) The effect of pH on the activity of lysozyme
In kinetics experiments using defined substrates, the E35Q and D52N mutants each showed <0.1% of wild-type activity, suggesting that both residues play critical roles in catalysis. As shown in Figure 11.15, lysozyme has optimum activity at ~5, and is dependent on a base with a $pK_{a}$ of ~4 and an acid with a $pK_{a}$ of ~6.
The $pK_{a} s$ of E35 and D52 were found to be, respectively, 6.2 and 3.7. Thus, at values of pH between 3.7 and 6.2, E35 would be predominantly protonated and D52 would be predominantly deprotonated, as required by the Phillips mechanism.
E35 must be protonated to act as a general acid catalyst in the first step of the mechanism; thus, at pH values below 6.2 the ratio of [COOH]/[COO^–^ ] is greatest, favoring catalysis.
D52 must be deprotonated to interact with the oxocarbenium ion; thus, at pH values above 3.7 the ratio of [COO^–^ ]/[COOH] is greatest, favoring catalysis.
These two boundary requirements give rise to the observed pH optimum (~5) where both protonated E35 and deprotonated D52 are abundant.
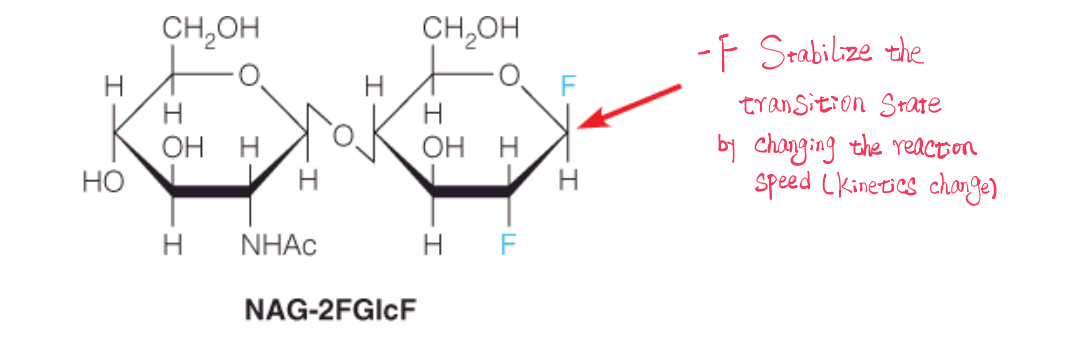
(4) Evidence for the covalent intermediate in the mechanism of lysozyme
The covalent adduct (加合物) between the synthetic substrate NAG-2FGlcF and D52 in the active site of the E35Q mutant of lysozyme is shown.
This substrate readily forms the oxocarbenium ion; however, the subsequent addition of -OH to the C1 is slowed by the E35Q mutation (Q does not act as a general base). Thus, the covalent intermediate is sufficiently long-lived that it is possible to obtain a crystal structure.
In summary, lysozyme employs GABC, substrate distortion, and covalent catalysis to achieve its rate enhancement.
Case 2: Chymotrypsin 胰凝乳蛋白酶,糜蛋白酶
serine proteases: the catalysis of peptide-bond hydrolysis
This important class of enzymes includes trypsin and chymotrypsin
The serine proteases are distinct because they all have a critical serine nucleophile in the active site.
A nucleophile is a chemical species that donates an electron pair to form a chemical bond in relation to a reaction. All molecules or ions with a free pair of electrons or at least one pi bond can act as nucleophiles. Because nucleophiles donate electrons, they are by definition Lewis bases.
(1) Catalysis of peptide bond hydrolysis by a serine protease
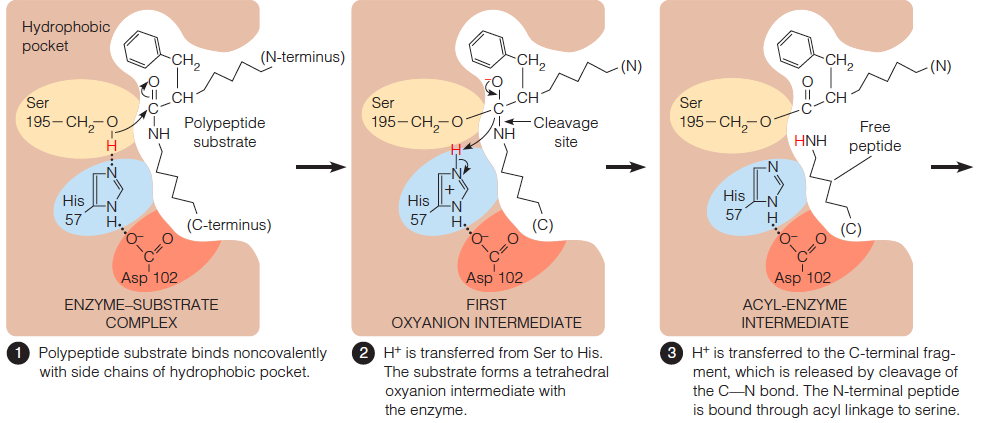
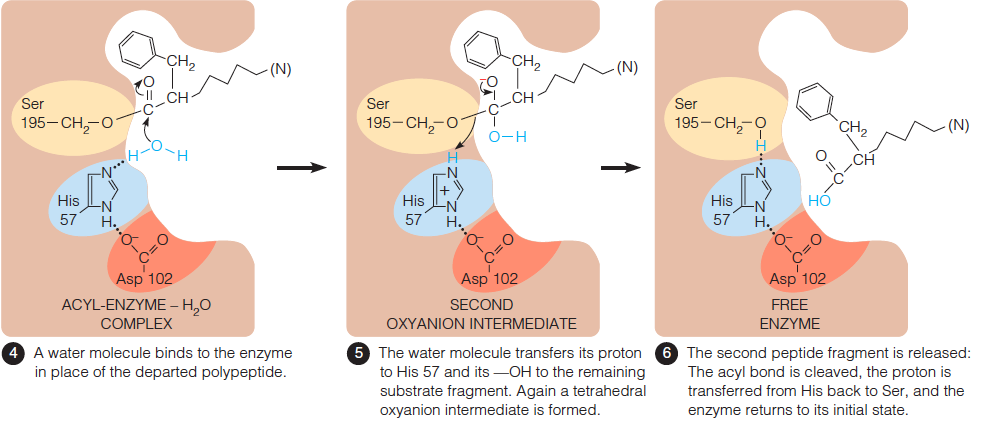
- First, the polypeptide chain to be cleaved is bound to the enzyme surface, Polypeptide substrate binds noncovalently with side chains of hydrophobic pocket.
- Most of the polypeptide binds non-specifically, but the side chain of the residue to the N-terminal side of the peptide bond to be cleaved must fit in the active site pocket.
- This pocket defines not only the position of the bond cleavage but also the specificity of serine proteases.
- Each of the serine proteases preferentially cleaves the amide bond immediately C-terminal to a specific kind of amino acid side chain.
- Second, $H^{+} $ is transferred from Ser to His. The substrate forms a tetrahedral oxyanion intermediate with the enzyme.
- $H^{+}$ is transferred to the C-terminal fragment, which is released by cleavage of the C–N bond. The N-terminal peptide is bound through acyl linkage to serine.
- A water molecule binds to the enzyme in place of the departed polypeptide.
- The water molecule transfers its proton to His57 and its –OH to the remaining substrate fragment. Again a tetrahedral oxyanion intermediate is formed.
- The second peptide fragment is released: The acyl bond is cleaved, the proton is transferred from His back to Ser, and the enzyme returns to its initial state.
( trypsin cleaves preferentially to the carboxylate side of basic amino acid residues like lysine or arginine, whereas chymotrypsin prefers a large hydrophobic residue like phenylalanine in this position. )
Trypsin, on the other hand, has an aspartate side chain in the bottom of the specificity pocket. This negatively charged residue provides complementary binding interactions to the positive charge on an arginine or lysine side chain.
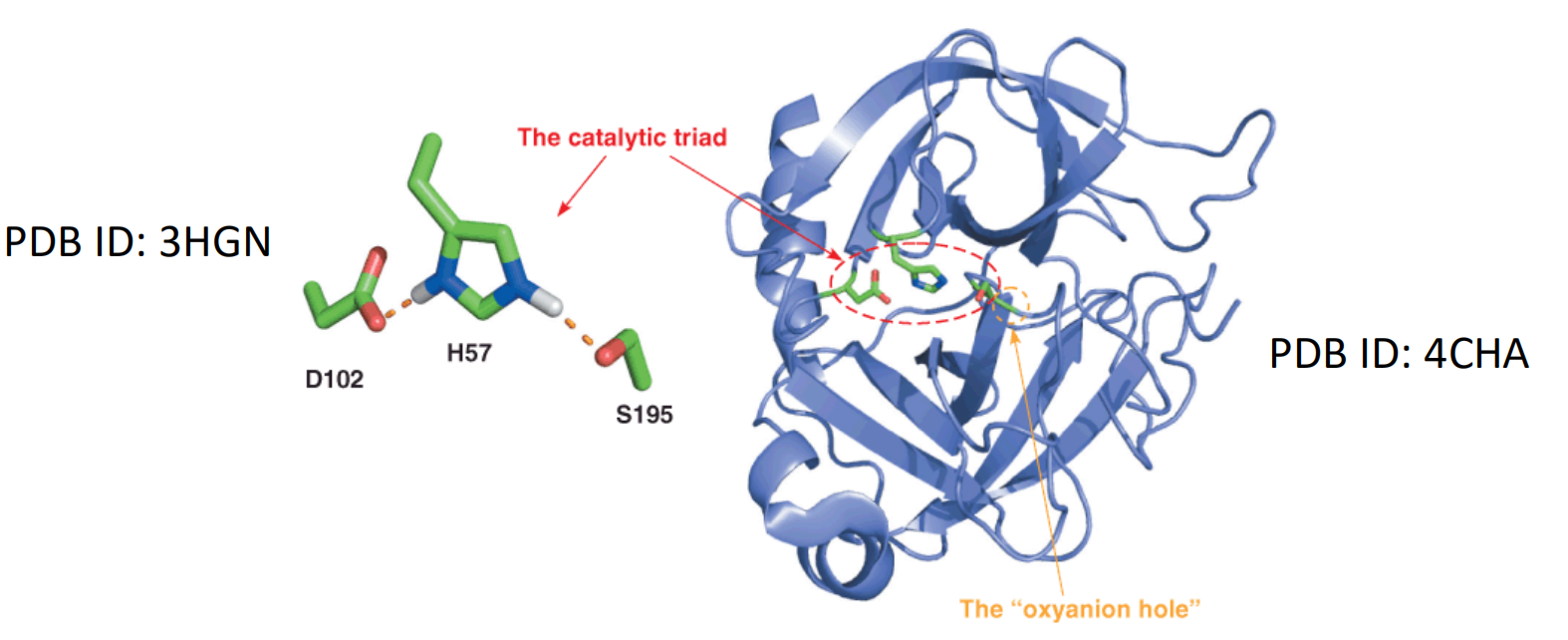
A common feature of serine proteases is the so-called catalytic triad of a nucleophile, a general base, and an acid.
Catalytic triad (催化三联体): nucleophile, a general base, an acid
催化三联体: 催化三联体通常指在水解酶和转移酶的活性位点中心同时作用的三个氨基酸残基(如蛋白酶、酰胺酶、酯酶、酰基转移酶、脂酶和β-内酰胺酶)。用于共价催化的亲核残基一般是酸-碱-亲核三联体。
(残基会形成一个电荷中继网络,以极化和活化亲核试剂,来进攻底物形成共价中间体,然后中间体水解,再生出游离的酶。亲核试剂大多是丝氨酸或半胱氨酸,也有少量是苏氨酸。)
In many of the serine proteases that have been studied in detail, this catalytic triad is composed of serine, histidine, and aspartic acid residues presented in a similar 3-D orientation in the active site.
In chymotrypsin the catalytic triad residues are Ser 195(S195), His 57 (H57), and Asp 102 (D102). Other serine proteases have been discovered in which the triad pattern nucleophile–base–acid is conserved but the identities of the base and acid vary (e.g., Ser–His–His or Ser–Glu–Asp).
(2) The structure of chymotrypsin and the serine protease catalytic triad:
The backbone of bovine chymotrypsin determined by X-ray crystallography.
Here, H57 is protonated, S195 and D102 are deprotonated.
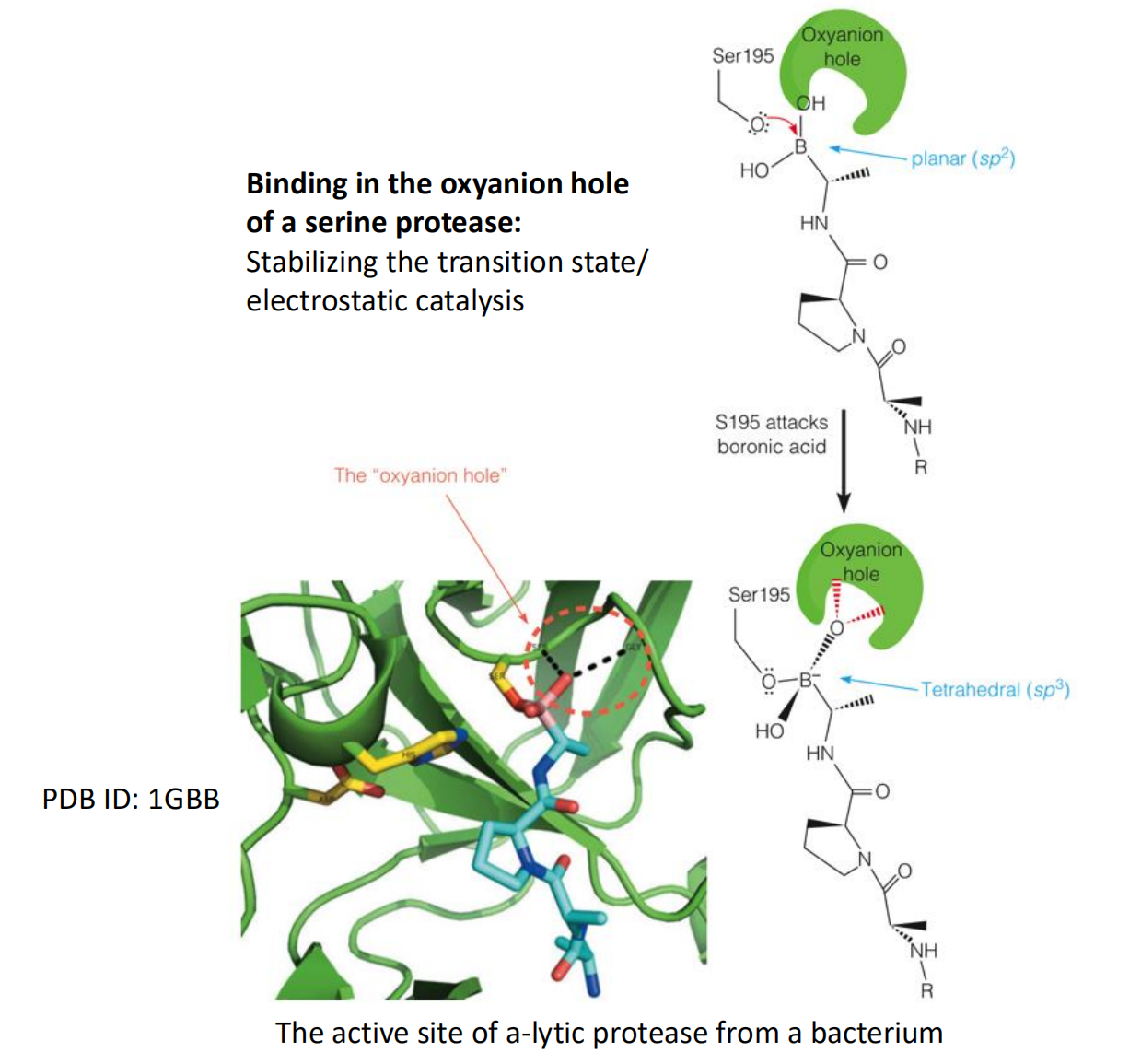
Enzymes Can Use Many Catalase Strategies Simultaneously

The role of Dynamics in Catalysis
Case 3: Dihydrofolate reductase 二氢叶酸还原酶
Dynamic conformational changes in the catalytic cycle
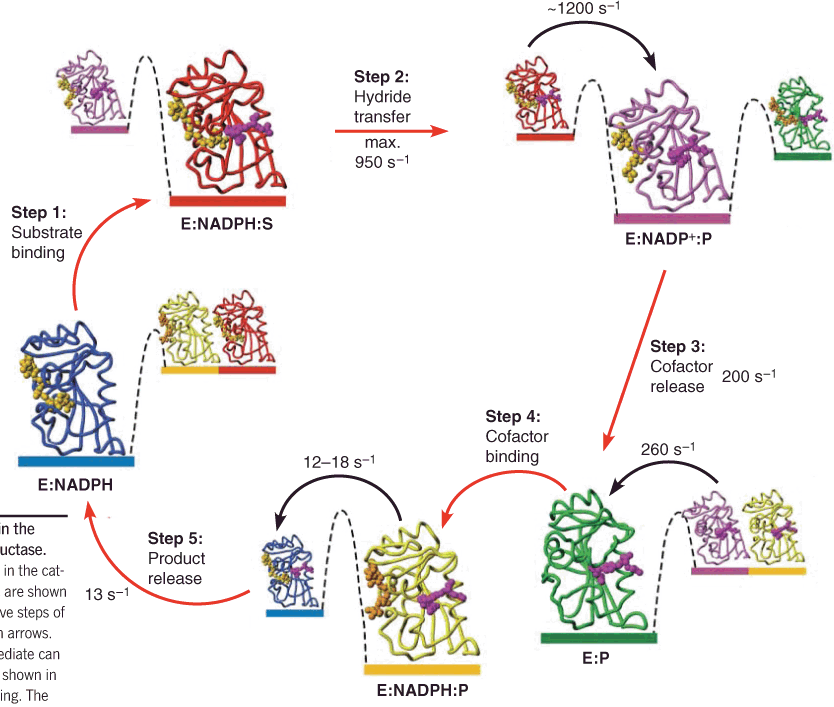
具有较低的构象均匀性(Conformation uniformity)/ conformational homogeneity (构象的同质性)
The difference between different stage is energy, but not the conformation of enzyme.
Dynamic conformational switching may require the correlated movements of several amino acid residues in the protein, and such concerted motions make up a dynamic “network.”
So:
- some mutations distant from the active site have significant impacts on reaction rates even though these amino acids make no direct contact to the substrate
- such mutations tend to affect the dynamic behavior of a larger group of amino acids.
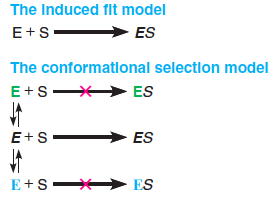
(1) Conformational Selection vs. Induced Fit:
Induced fit implies conformational homogeneity (构象的同质性) in the unbound enzyme, which is then distorted upon substrate binding.
Conformational selection implies that the unbound enzyme is present in multiple conformations; but, the substrate can only bind to unbound enzyme that is in the same conformation as that of the ES complex.
Substrate binding will perturb (使(某人)烦恼;不安) the conformational equilibrium of the unbound enzyme, driving more E into the ES conformation by Le Chatelier’s principle.