L6 Cell membrane proteins add functionality to cell membrane
一、Membranes
Membrane protein functions
membrane proteins are amphiphilic, having hydrophobic and hydrophilic regions
1. Selectively Permeable
Based on the molecule size and charge
steroids are large molecule but highly hydrophobic
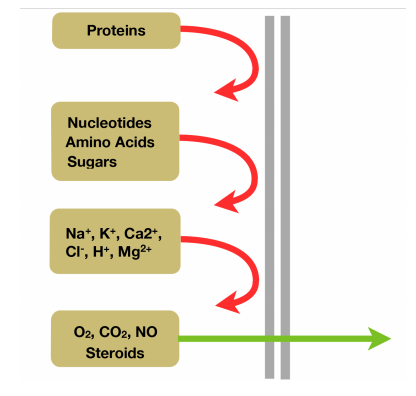
2. Sense Surrounding Environment
3. Help Cells Stay Together
4. Other Functions
Cell membrane contains proteins for transport of materials, adhesion and detection
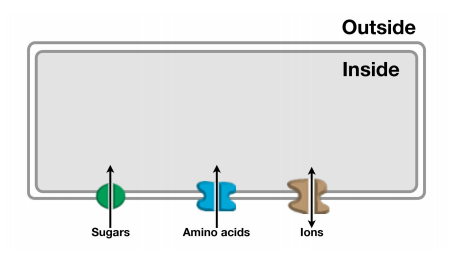
Protein channels allow passage of small molecules and ions across membranes
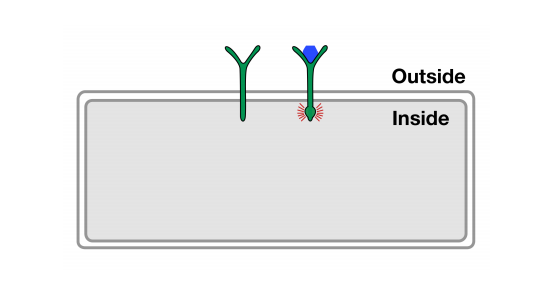
Receptors allow cells to sense and communicate with outside world
Adhesion proteins in the cell membrane hold cells together
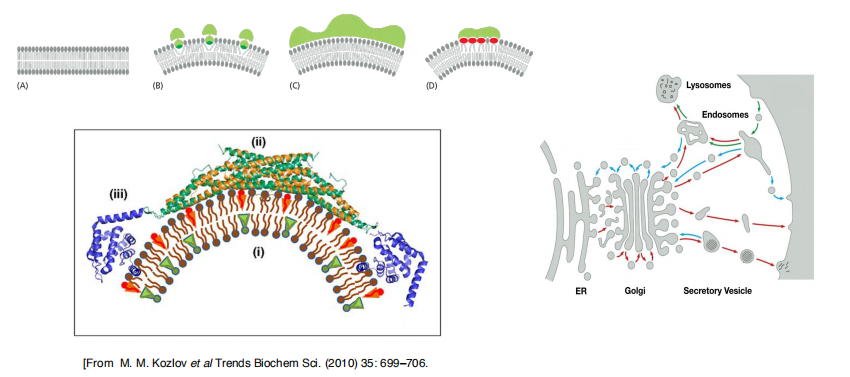
Membrane-bending proteins deform bilayer (contribute to membrane curvature)
- Some insert hydrophobic protein domains or attached lipid anchors into one of the leaflets of a lipid bilayer. Increasing the area of only one leaflet causes the membrane to bend. The proteins that shape the convoluted network of narrow ER tubules are thought to work in this way
- Some membrane-bending proteins form rigid scaffolds that deform the membrane or stabilize an already bent membrane . The coat proteins that shape the budding vesicles in intracellular transport fall into this class.
- Some membrane-bending proteins cause particular membrane lipids to cluster together, thereby inducing membrane curvature.
- The ability of a lipid to induce positive or negative membrane curvature is determined by the relative cross-sectional areas of its head group and its hydrocarbon tails. For example, the large head group of phosphoinositides make these lipid molecules wedge-shaped, and their accumulation in a domain of one leaflet of a bilayer therefore induces positive curvature (Figure 10–40D). By contrast, phospholipases that remove lipid head groups produce inversely shaped lipid molecules that induce negative curvature.
5. Proteins add functionality to cell membrane
Membrane proteins fulfill diverse functions:
- Compartmentalization/border between cell & environment $\rarr$ that is why it has to be a relatively impermeable barrier
- Gatekeeper, transport across the membrane, highly selective $\rarr$ selectively let some materials in and let others out)
- Receptors senses extracellular signals (signal perception/ligand binding)
- Cell-cell recognition
- Enzymatic activity (location-specific)
- Energy transduction (establish ion gradients to drive ATP synthesis or produce and transmit electric signals)
- Scaffolding
- Intercellular junctions
- attachment of cytoskeletal elements and/or extracellular matrix attachment $\rarr$ that’s why they are important therapeutic(治疗剂) targets
There are over 17,000 structures of water-soluble proteins, but only ~150 unique structures of membrane proteins.
Structural analysis of membrane proteins is extremely difficult. Why?
Categories of membrane proteins
membrane-associated proteins do not extend into the hydrophobic interior of the lipid bilayer at all; they are instead bound to either face of the membrane by noncovalent interactions with other membrane proteins.
Many of the proteins of this type can be released from the membrane by relatively gentle extraction procedures, such as exposure to solutions of very high or low ionic strength or of extreme pH, which interfere with protein–protein interactions but leave the lipid bilayer intact; these proteins are often referred to as peripheral membrane proteins.
1. Membrane proteins: functional groups
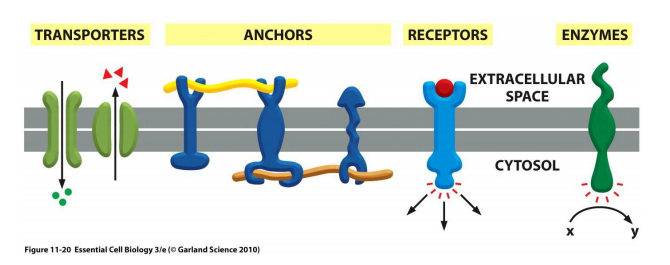
- Transporters: e.g. Na+ pump (ion channels)
- Anchors: e.g. integrins (Help the cell-ECM(Extracellular Matrix) interaction, the cell in this way anchored to the ECM)
- Receptors: e.g. EGF (epithelial growth factor) receptor
- Enzymes: e.g. adenylyl cyclase (catalyze cAMP production)
2. Membrane proteins: positional groups
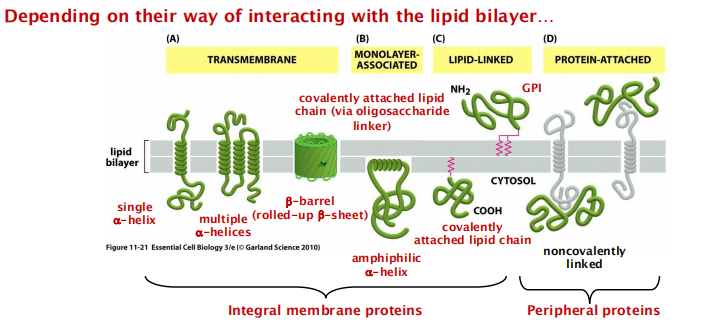
Some positional groups:
Transmembrane (integral, IMP)
Monolayer-associated (IMP)
Lipid-linked (IMP)
Protein-attached (peripheral, PMP)
Integral and peripheral membrane proteins
There are different types of membrane proteins and they are classified according to the way how they are associated with the lipid bilayer:
Integral membrane protein
Transmembrane proteins: possess transmembrane domain(s)
Covalently linked proteins: linkage to lipid groups (also via oligosaccharides) or insertion of hydrophobic regions into the lipid bilayer)
Peripheral membrane proteins
- only associated with the membrane or noncovalently linked with other membrane proteins
(1) Integral Membrane Proteins
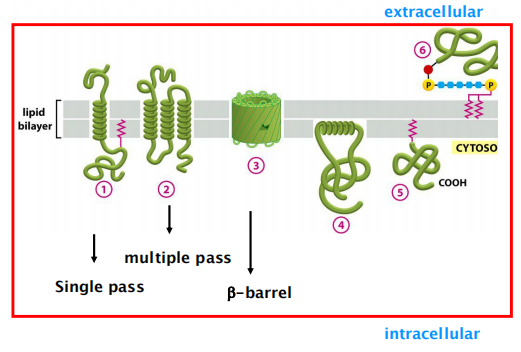
Penetrate the bilayer or span the membrane entirely.
Two types of integral membrane proteins:
- transmembrane proteins.
- covalently tethered protein:
- covalently linked to phospholipids or to glycolipids.
Many integral proteins are glycosylated (糖基化的).
- The proteins are covalently linked via Asn (asparagine), Ser (serine), or Thr (threonine) residues to sugars
Most transmembrane proteins in animal cells are glycosylated. As in glycolipids, the sugar residues are added in the lumen of the ER and the Golgi apparatus
- the oligosaccharide chains are always present on the non-cytosolic side of the membrane.
This environment decreases the likelihood that intrachain or interchain disulfide (S–S) bonds will form between cysteines on the cytosolic side of membranes.
These bonds form on the non-cytosolic side, where they can help stabilize either the folded structure of the polypeptide chain or its association with other polypeptide chains
they can help stabilize either the folded structure of the polypeptide chain or its association with other polypeptide chains
Because the extracellular part of most plasma membrane proteins are glycosylated, carbohydrates extensively coat the surface of all eukaryotic cells.
- Proteoglycans (蛋白聚糖), which consist of long polysaccharide chains linked covalently to a protein core, are found mainly outside the cell, as part of the extracellular matrix
- The terms cell coat or glycocalyx(糖被) are sometimes used to describe the carbohydrate-rich zone on the cell surface.
- One of the many functions of the carbohydrate layer is to protect cells against mechanical and chemical damage; it also keeps various other cells at a distance, preventing unwanted cell–cell interactions.
The topology of transmembrane proteins is diverse.
They can only be removed from membranes by disrupting the lipid bilayer.
(2) Peripheral membrane proteins
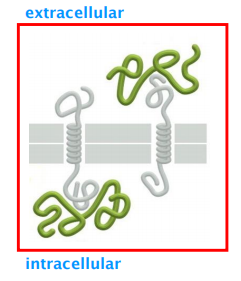
Characteristics:
- Do NOT penetrate the lipid bilayer and do NOT covalently link to other membrane components, but do form ionic links to membrane structures.
- Dissociation of peripheral membrane proteins with chaotropic(离液序列高的) agents does not disrupt membrane integrity.
- Can locate to both, extracellular and intracellular side of the membrane
- Often link membrane to non-membrane structures.
Synthesis of peripheral proteins for the plasma membrane (PM) occurs at different locations, dependent on which side of the PM they attach:
- attachment at cytoplasmic (inner) side: synthesized in the cytoplasm.
- attachment at extracellular (outer) side: synthesized in ER and are then delivered to the extracellular side by exocytosis/secretion.
Experimentally test whether a membrane protein is integral or peripheral
Integral membrane proteins are NOT released by harsh salt concentrations or extreme pH, which would change the ionic interactions between proteins or protein/polar groups of lipids
Peripheral membrane proteins are released by harsh salt concentrations or extreme pH.
Structural characteristics of transmembrane proteins
1. Structural characteristics of proteins associated with the membrane
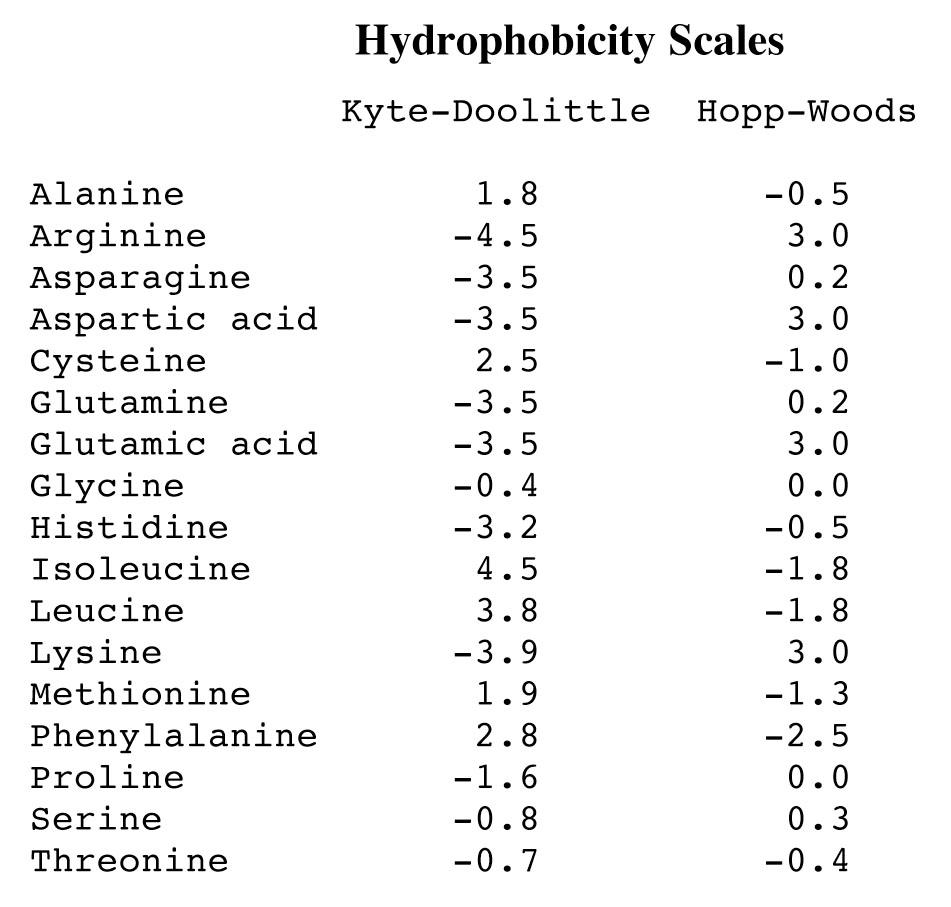
Hydrophobicity prediction based on the amino acid sequence.
左:Hydrophobicity scale; 右:Hydrophilicity Scale
Membrane proteins : secondary structure
- Mostly $\alpha$-helices
- Sometimes also $\beta$-barrels (e. g. porins)
Most transmembrane proteins crosses the lipid bilayer in $\alpha$-helical conformation
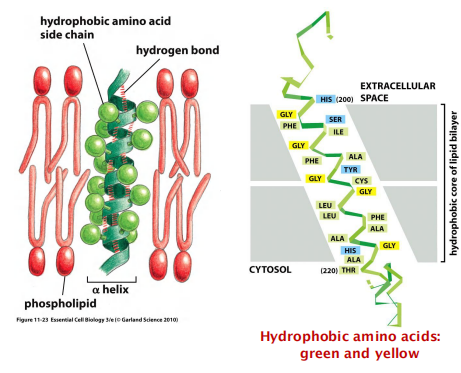
Transmembrane $\alpha$-helices
A segment of 20-30 amino acids commonly with a high degree of hydrophobicity
Hydrophobic amino acids are outside, facing the hydrophobic fatty-acid chains of the membrane
Localizing $\alpha$-helical transmembrane segment with hydropathy
Hydropathy plots to predict transmembrane $\alpha $-helices this is important for the topology prediction of a transmembrane protein
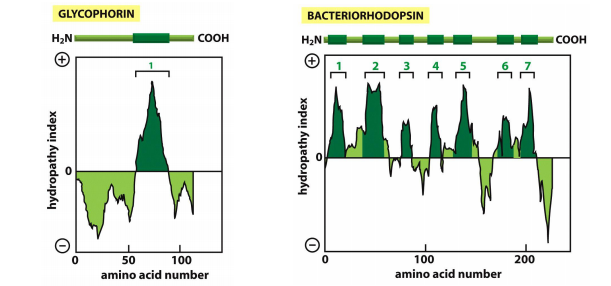
Hydrophopathy index:
- Positive value indicates free energy is required for transfer to water
- More positive indicates more hydrophobic
2. Not all membrane interacting regions can be predicted
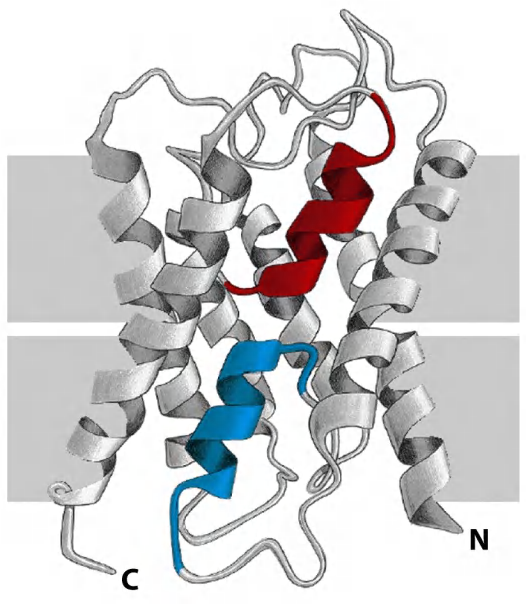
Some transmembrane protein regions can’t be predicted by hydropathy plots, these includes
The β-barrels, as they are short and only every other amino acids is hydrophobic.
Membrane proteins which do not contact hydrophobic bilayer, but rather interact with other transmembrane proteins.
- Colored two α-helices in an aquaporin water channel are buried at an interface formed by protein-protein interactions, they are not hydrophobic.
3. Folding of multipass transmembrane proteins
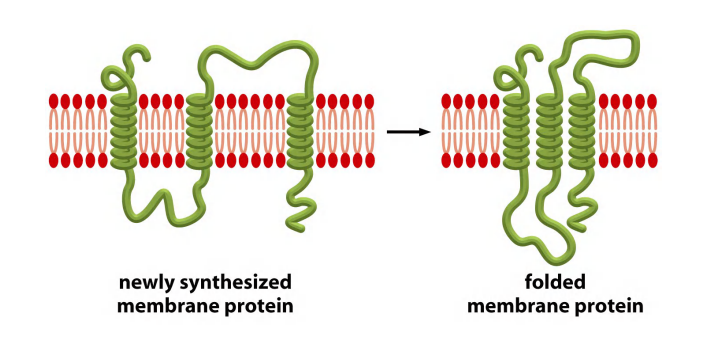
Transmembrane $\alpha$-helices often interacts with one another
A transmembrane $\alpha $-helix has specificity for its interaction partners
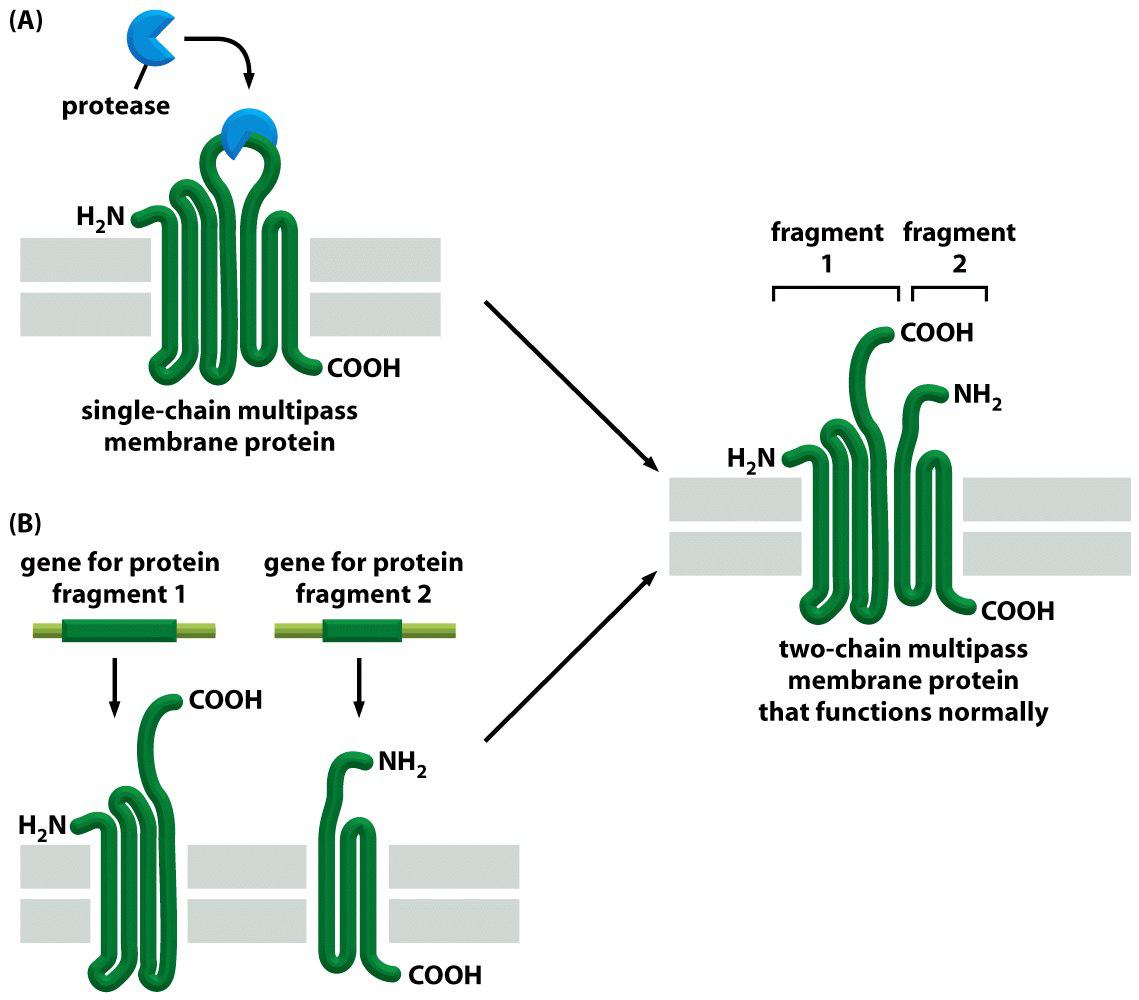
Multiple pass transmembrane protein with 6 $\alpha$-helices
Example 1: mitochondrial ATP/ADP carrier protein (multiple pass transmembrane protein)
6 $\alpha $-helices form a compact transmembrane domain
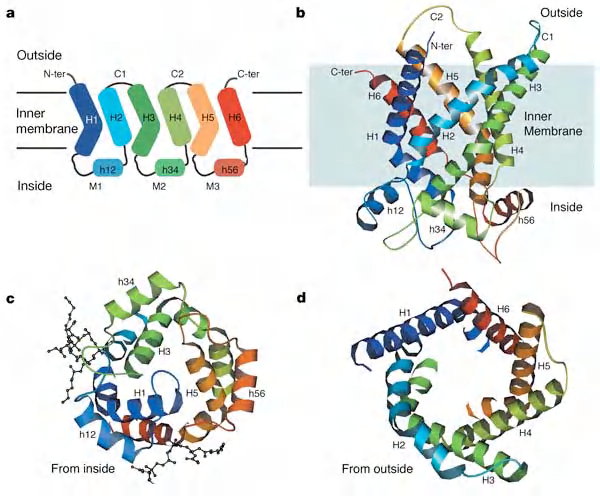
Bacteriorhodopsin
Each bacteriorhodopsin molecule is folded into seven closely packed trans-membrane α helices and contains a single light-absorbing group, or chromophore
Bacteriorhodopsin: Multiple pass transmembrane protein with 7 $\alpha $-helices
- Light-driven proton pump: the first membrane protein whose structure was determined.
- Exist in the plasma membrane of the archaea Halobacterium salinarum
Pumps H+ in the sunlight and builds proton gradients across the membrane.
- changes conformation in response to light
Uses the H+ gradients to synthesize ATP or to drive other energy requiring activities.
Bacteriorhodopsin:
- 3 monomers
- Each monomer has 7 transmembrane Domains (TMDs)
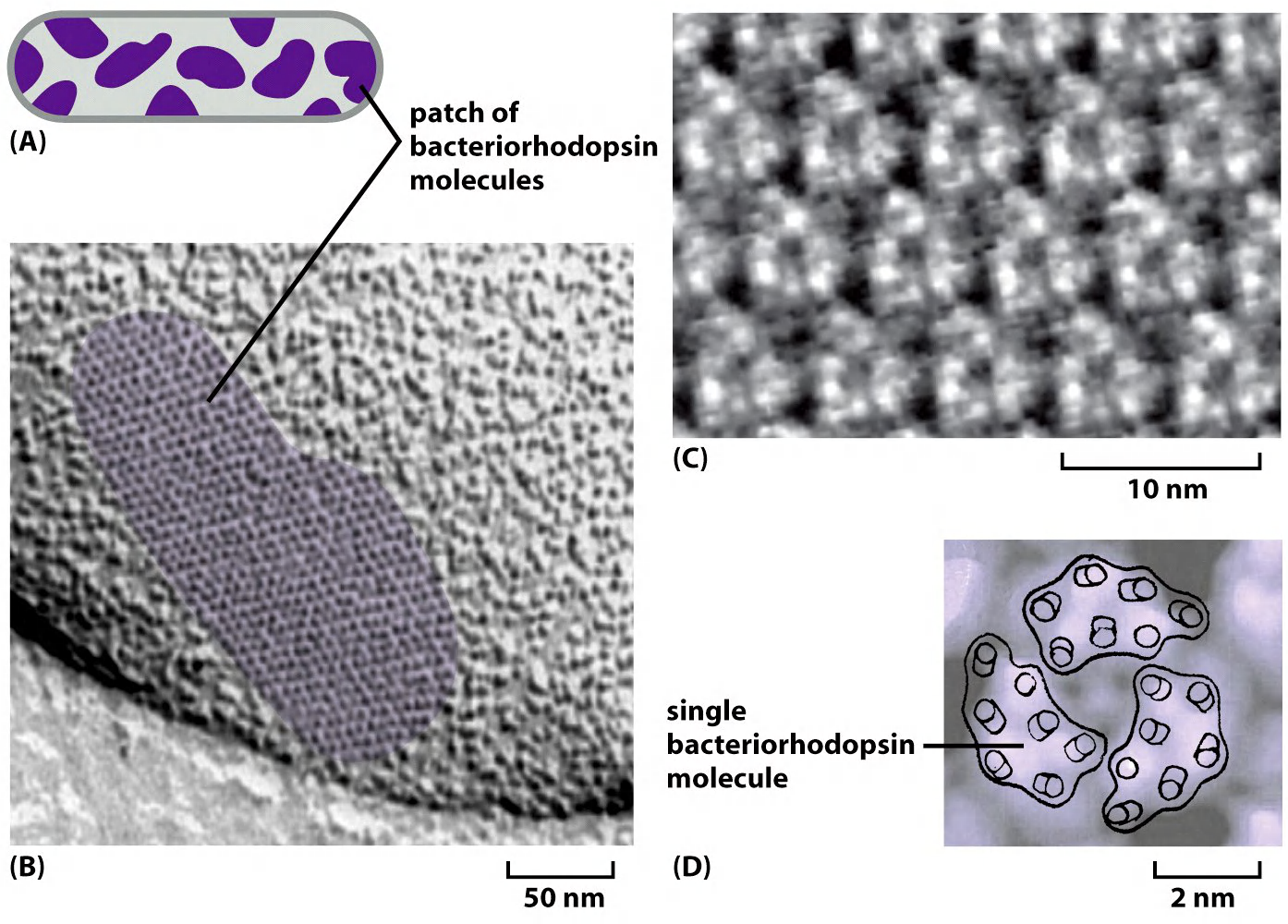
Small protein of ≈ 250 amino acids
Proton pump activated by light (pump proton out of the cell)
(1) 3-D structure of bacteriorhodopsin
Bacteriorhodopsin: light-driven proton pump
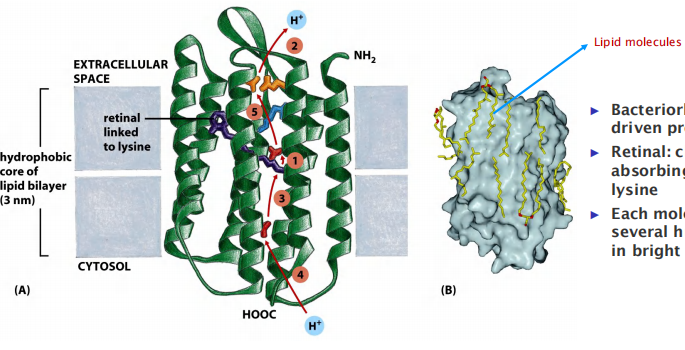
Retinal(视网膜的): chromophore (light absorbing group) – linked to a lysine
Each molecule can pump several hundred H+ per second in bright light
4. Transmembrane channels formed by $\beta $-barrel proteins
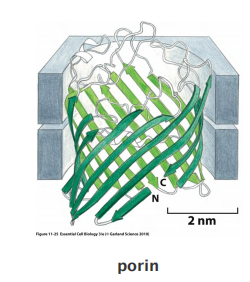
multiple transmembrane strands of a polypeptide chain to be arranged as a β sheet that is rolled up into a cylinder (a so-called β barrel;
Progress in the x-ray crystallography of membrane proteins has enabled the determination of the three-dimensional structure of many of them.
Hydropathy plots cannot identify the membrane-spanning segments of a β barrel, as 10 amino acids or fewer are sufficient to traverse a lipid bilayer as an extended β strand and only every other amino acid side chain is hydrophobic.
multi-pass transmembrane proteins can also contain regions that fold into the membrane from either side, squeezing into spaces between transmembrane α helices without contacting the hydrophobic core of the lipid bilayer.
Because such regions interact only with other polypeptide regions, they do not need to maximize hydrogen-bonding; they can therefore have a variety of secondary structures, including helices that extend only part way
Relatively rigid structures, conformational changes are less likely to occur.
Inside barrel: polar amino acids.
Outside barrel: nonpolar amino acids.
Abundant in outer membranes of mitochondria, chloroplast and bacteria.
Most $\beta $-barrel proteins have a “transporting function”
- e.g. porins but there are also are receptors and enzymes.
- Loops of the polypeptide chains protruding into the lumen of the channel confer selectivity: only selected molecules can pass.
Different types of $\beta$-barrel proteins
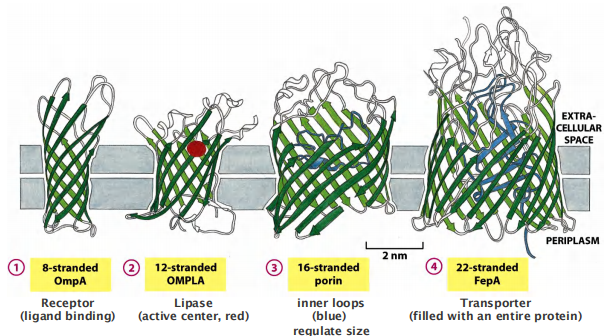
Some $\beta$-barrels form large Transmembrane Channels:
Multi-pass membrane proteins that have their transmembrane segments arranged as β barrels rather than as α helices are comparatively rigid and therefore tend to form crystals readily when isolated
Many porin barrels are formed from a 16-strand, antiparallel β sheet rolled up into a cylindrical structure
Found in: outer membrane of mitochondria and plastids
Not all β-barrel proteins are transport proteins. Some form smaller barrels that are completely filled by amino acid side chains that project into the center of the barrel. These proteins function as receptors or enzymes
Number of $\beta $-sheets: 8-22
In β-barrel proteins, by contrast, hydrogen bonds bind each β strand rigidly to its neighbors, making conformational changes within the wall of the barrel unlikely.
Synthesis of Integral membrane proteins
1. Synthesis of integral membrane proteins
Occurs in the (rough) endoplasmic reticulum (ER)
Many integral membrane proteins are glycoproteins:
- glycosylation (糖基化) begins in lumen of ER (for details, please referred to lecture 10)
- carbohydrates are modified in Golgi apparatus
Lipid–linked proteins are firstly made in cytosol as soluble proteins; they become a membrane protein after the linkage to the lipids
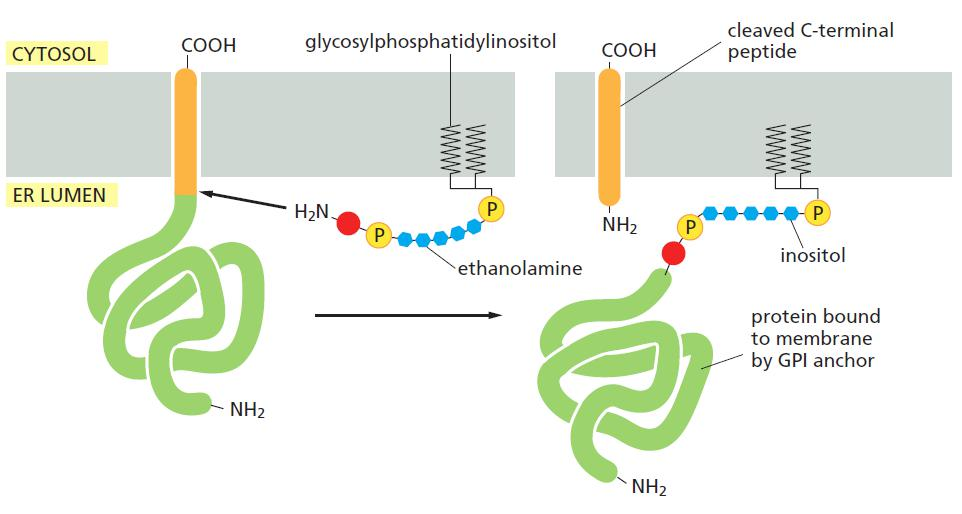
Glycosylphosphatidylinositol (GPI,糖基磷脂酰基醇) anchors are made as transmembrane proteins, after cleavage of transmembrane domain, it is linked by GPI anchor and targeted to membrane.
2. Post-translational modification of membrane proteins
Oligosaccharide chains(寡糖链) are diverse and are always on the exoplasmic side of plasma membrane proteins.
Due to reducing environment of the cytosol, disulfide bonds form only very rarely
Disulfide bonds on membrane proteins form extensively in the oxidizing environment in the ER lumen and are therefore mainly found on the exoplasmic side.
Disulfide bonds help to stabilize/determine the structure of proteins
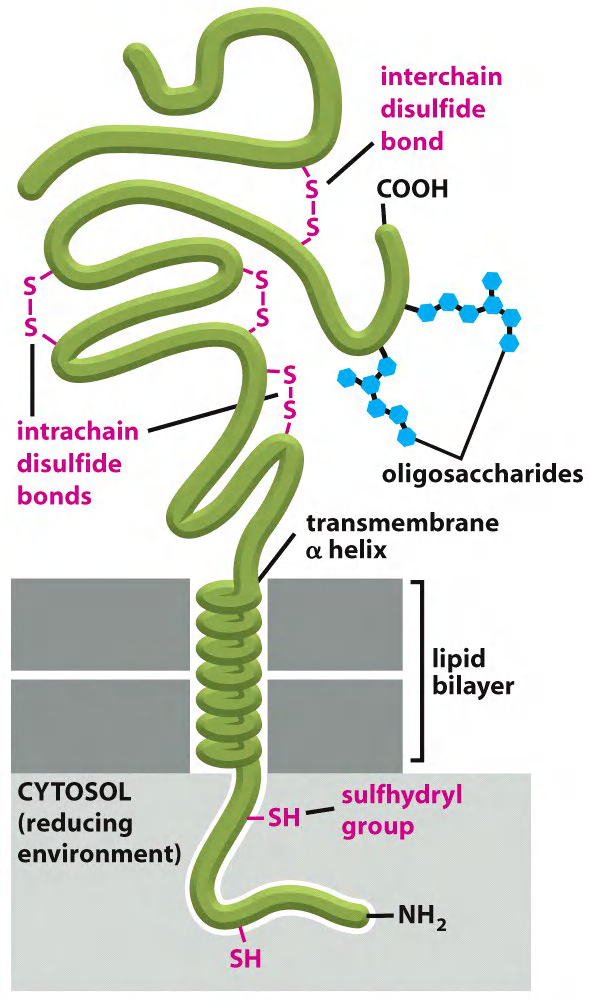
Glycosylation & disulfide bridges: always on the cell surface
Glycosylphosphatidylinositol (GPI)-anchored Proteins
GPI-anchored proteins are synthesized as integral membrane proteins in the ER, and are post-translationally processed (cleavage and transfer to the anchor).
Proteoglycan 蛋白聚糖
- part of extracellular matrix
- long polysaccharide chains(多聚糖链) linked covalently to a proteisn core
- integral proteins
- GPI-anchored proteins
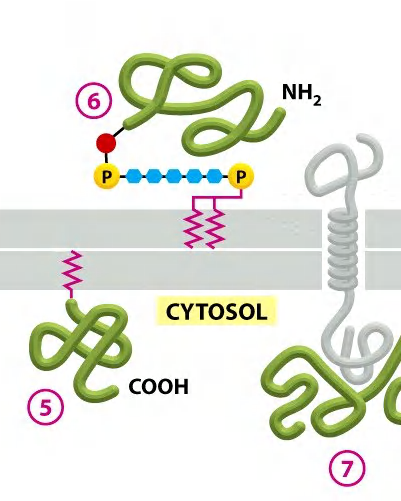
How to discriminate between glycosylated transmembrane proteins and GPI-anchored proteins?
Digestion using phosphatidylinositol-specific phospholipase C
GPI anchored protein
- can be recognized by phosphatidylinositol (PI)-specific phospholipase C
- can be cleaved off from the membrane
3. Covalent attachment of membrane proteins to lipids
Different ways for membrane proteins to covalently attach to membrane lipid.
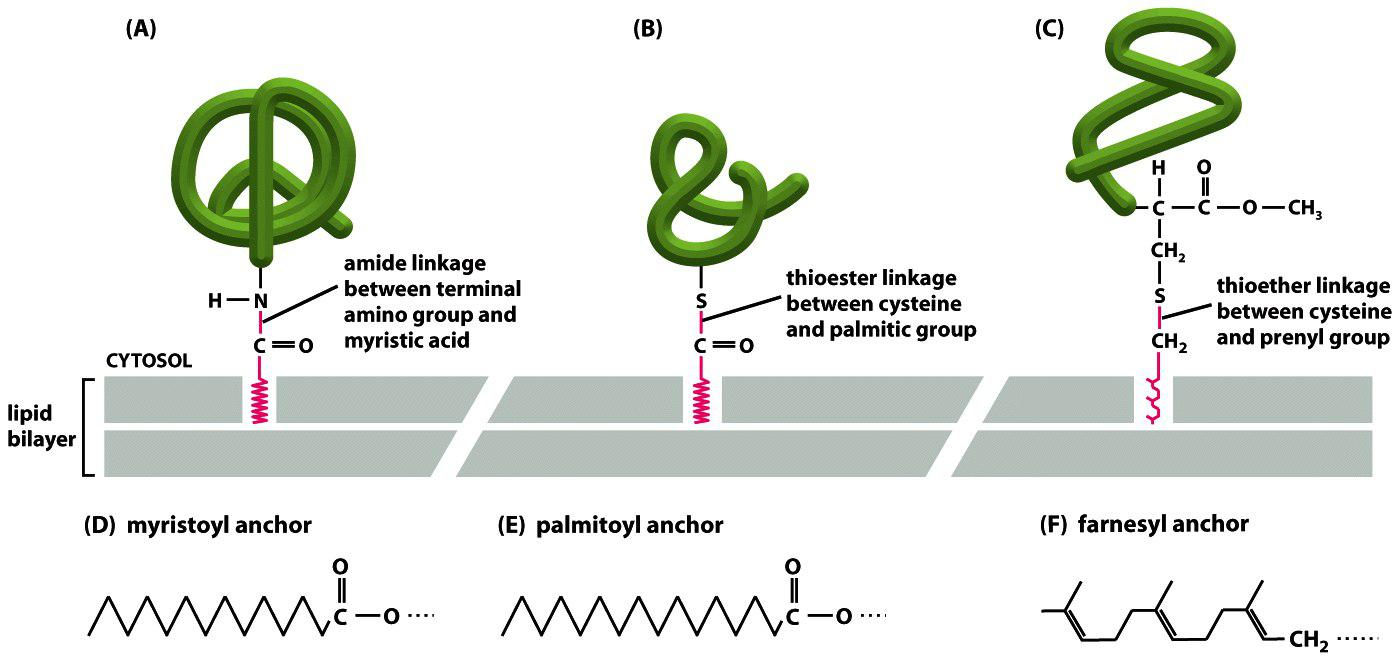

Lipid anchors span only one layer of the bilayer!
Mobility of membrane proteins
The fusion of proteins in the plasma membrane:
Membrane proteins move in the lipid bilayer
- Rotational mobility 旋转运动
- Lateral diffusion 横向扩散
- Protein mobility varies greatly:
- Some proteins are free to move
- Others are restricted in mobility
1. Plasma membrane domains
Some proteins have restricted mobility. This allows for specific accumulation of proteins in specific domains within a membrane!
Example: epithelium cells tight junction
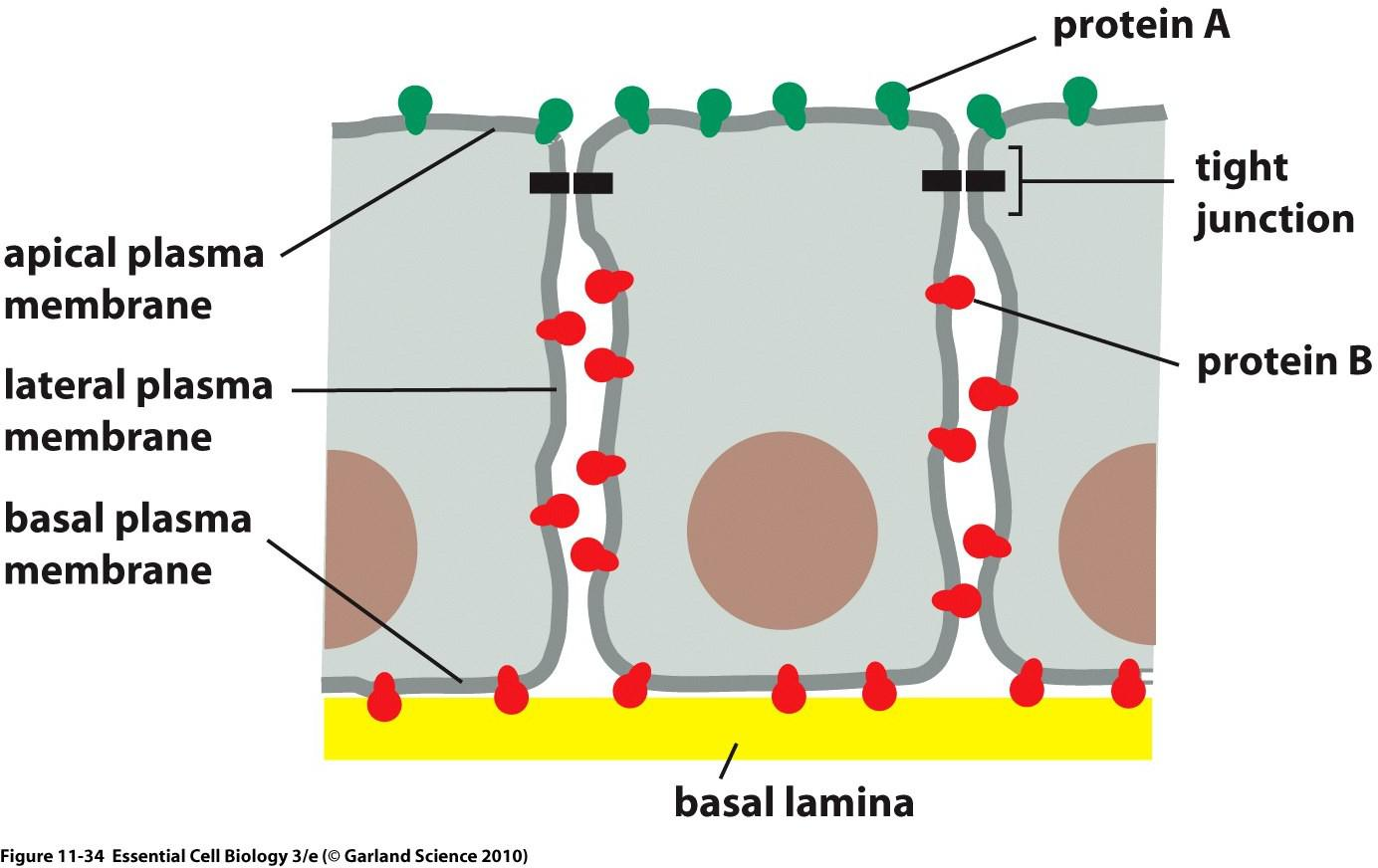
Tight Junctions: hold cells together and prevent diffusion of membrane proteins AND lipids to maintain distribution.
2. Three domains in the plasma membrane of sperm cells
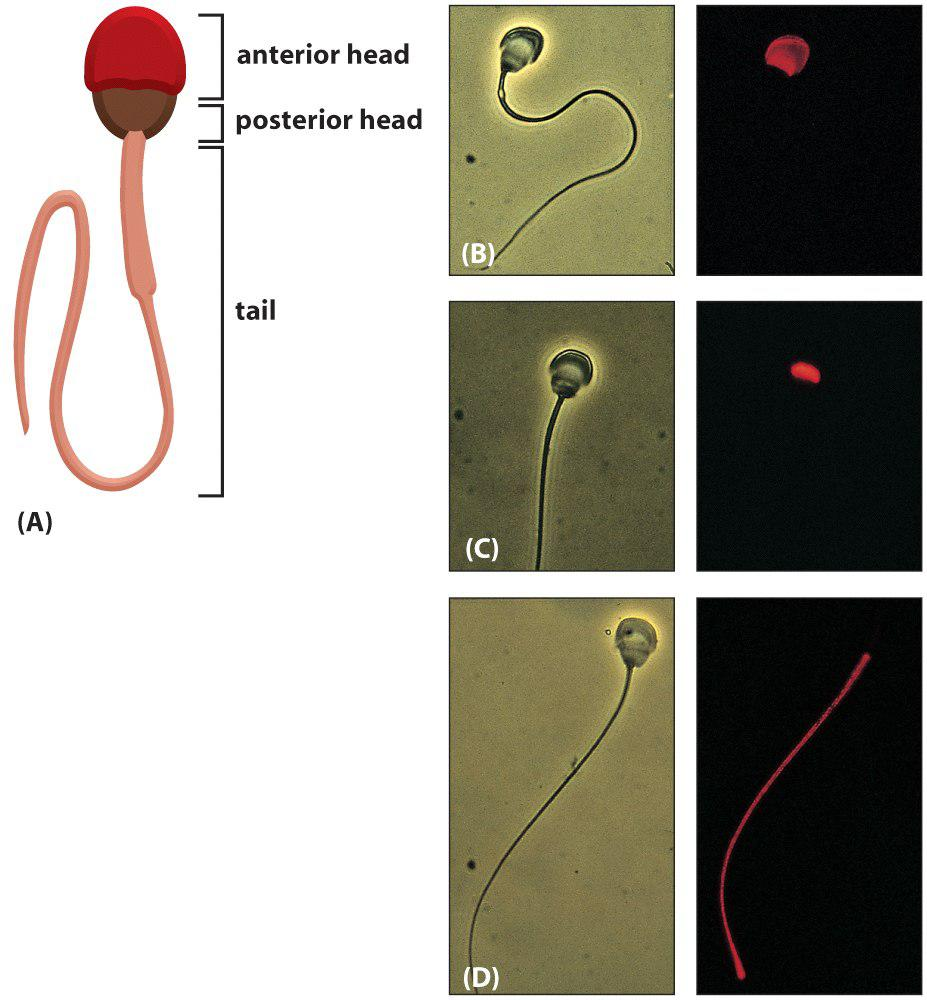
The plasma membrane of a guinea pig sperm cell contains continuous plasma membrane with three different domains.
- Mechanism: not clear yet!
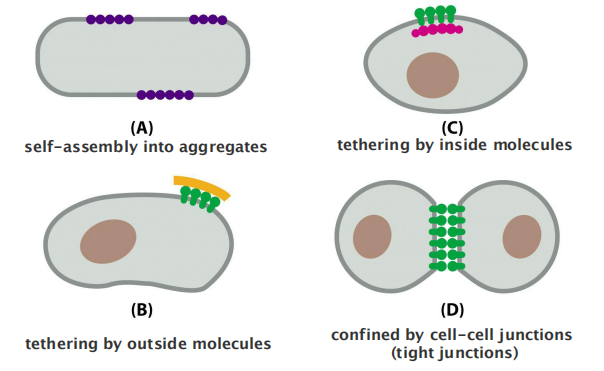
How can membrane proteins form/accumulate in domains?
4 different ways to accumulate membrane proteins in a domain:
- self-assembly into aggregates
- tethering by inside molecules
- tethering by outside molecules
- confined by cell-cell junctions (tight junctions)
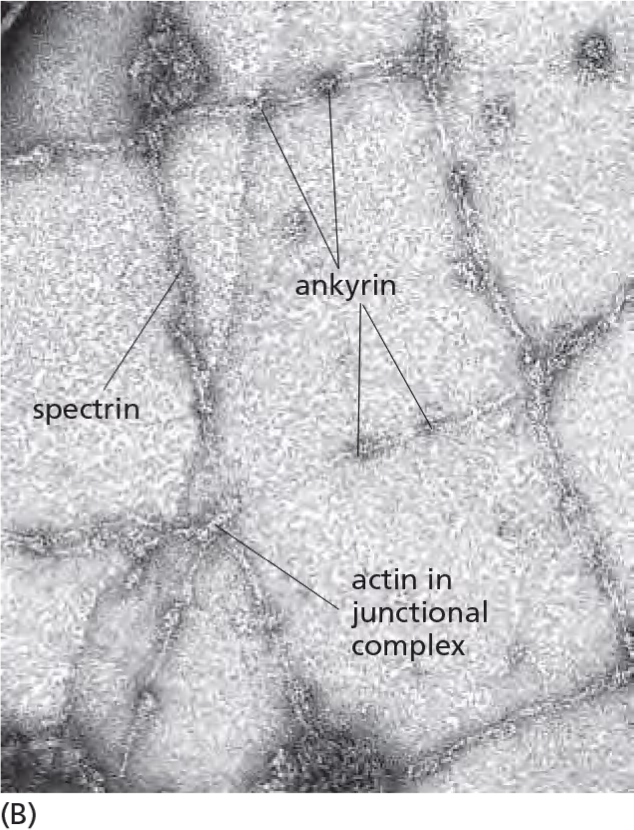
3. Spectrins: cytoskeleton network restricts membrane protein diffusion
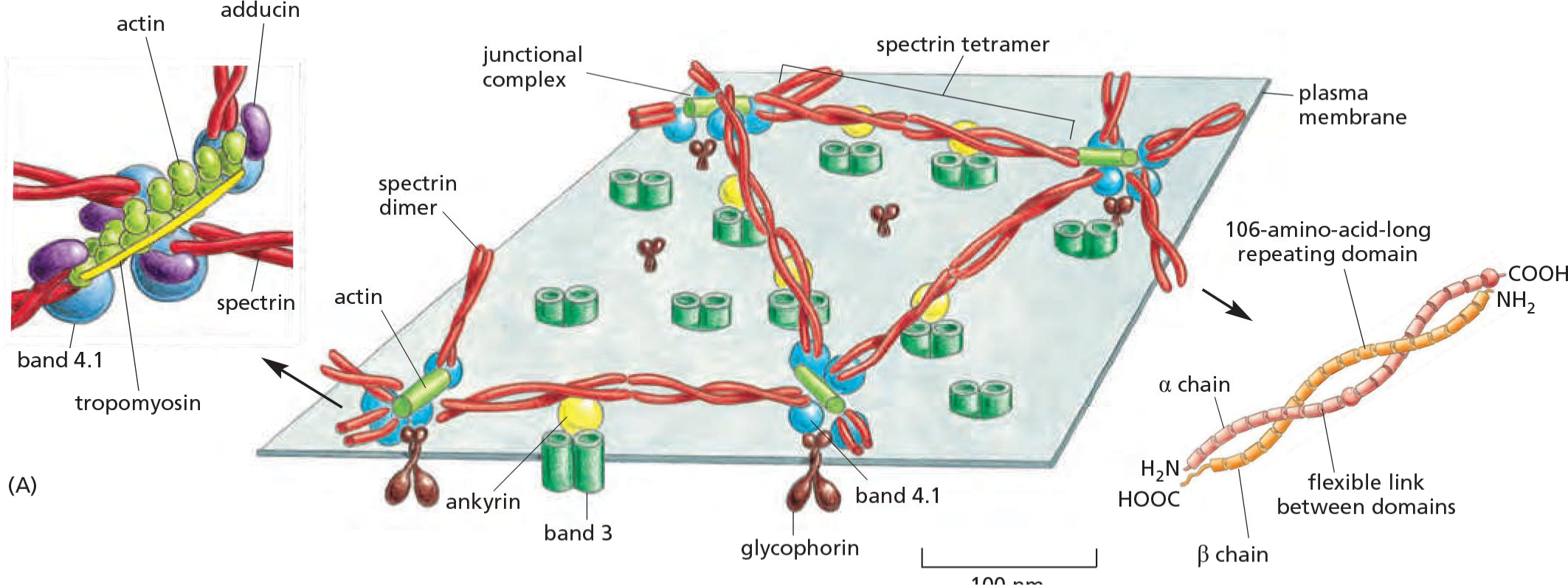
Spectrin (血影蛋白) network (red) interacts with a junctional complex in red blood cells:
- Results: Shaping of cells: the characteristic shape of the red blood cell.
- Retention works best if membrane proteins have large cytosolic domains which many PM proteins have!
4. Corralling of membrane proteins by cortical cytoskeletal filaments
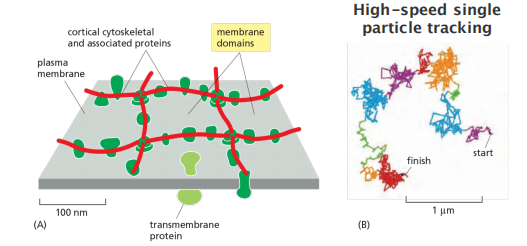
Membrane proteins can be kept in place by the cytoskeleton: “Corralling”. (corral,畜栏)
The attached cytoskeleton acts as a grid(格子,栅栏) that prevents lateral diffusion!
The extent to which a transmembrane protein is confined within a corral depends on its association with other proteins and the size of its cytoplasmic domain.
How can cytosolic domains become so large?
- To stay at specific domain.
5. Distribution of membrane proteins in the membrane
Membrane protein are asymmetrically distributed
- Each membrane protein has a unique conformation and orientation (Conformational changes of protein can frequently occur but that depends on protein function)
- Flip-flop (change between the leaflets) of proteins does NOT occur
- Proteins can be confined to specific membrane domains
- Carbohydrates of glycoproteins are always at the outer surface
- Disulfide bonds are also always at the outer surface
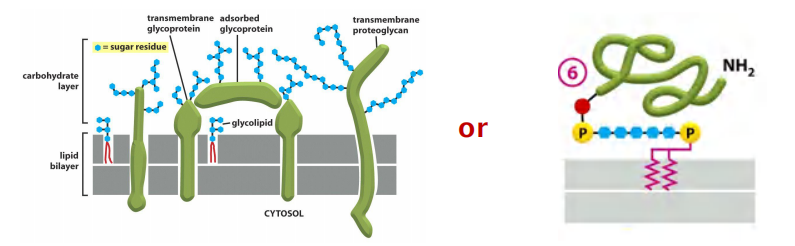
Carbohydrate layer
The oligosaccharide layer consists of glycoproteins & glycolipids.
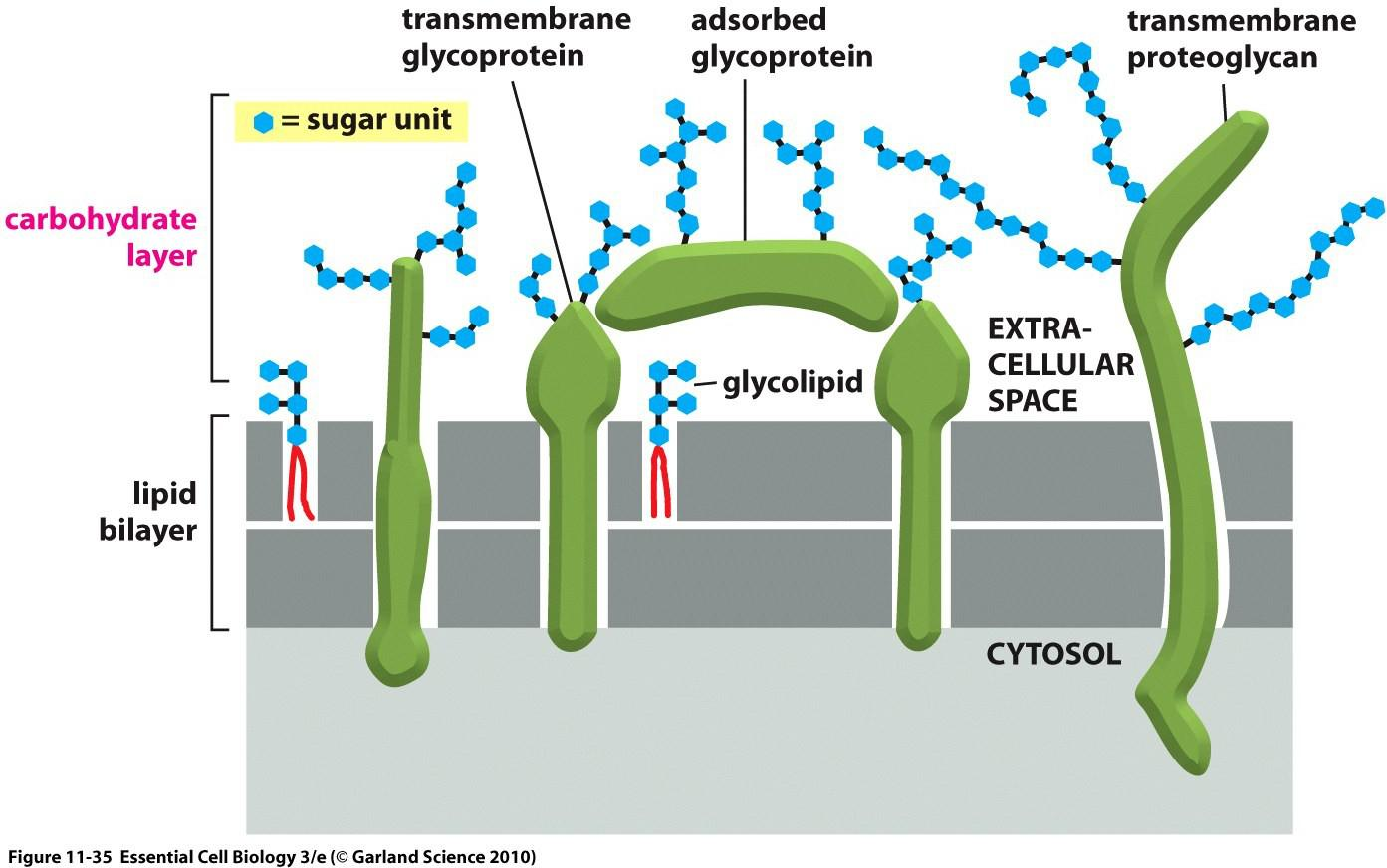
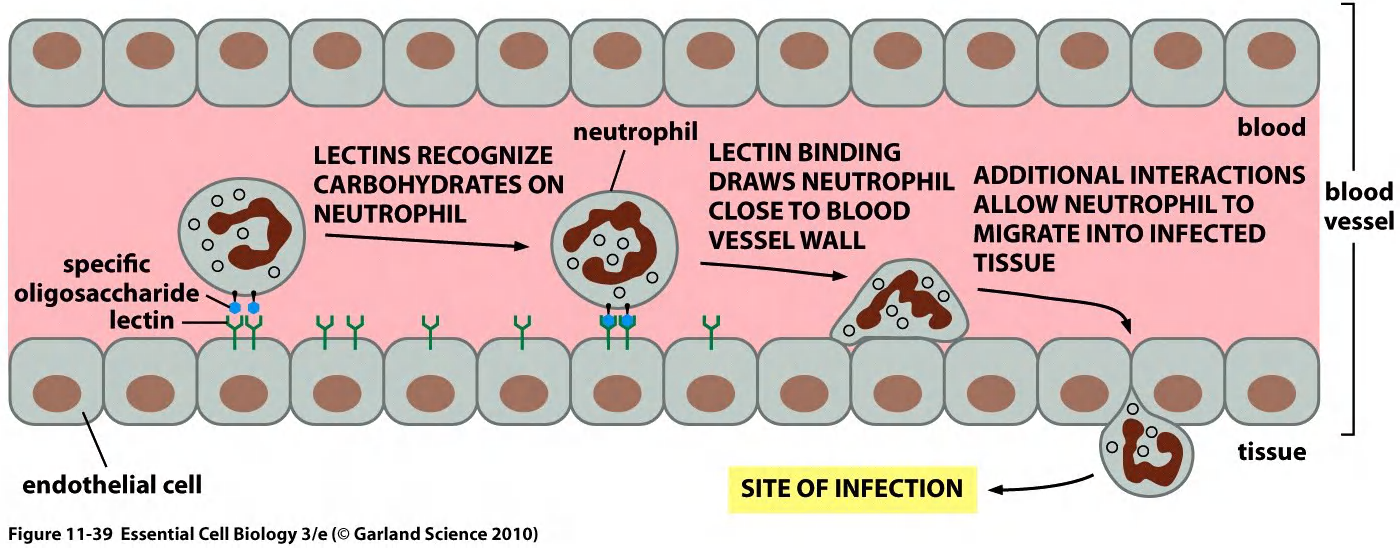
Glycoproteins can be specifically recognized by carbohydrate-binding proteins, termed lectins(凝集素). $\rarr $ fluorescently labeled lectins carbohydrate-binding can be used to label carbohydrate layer.
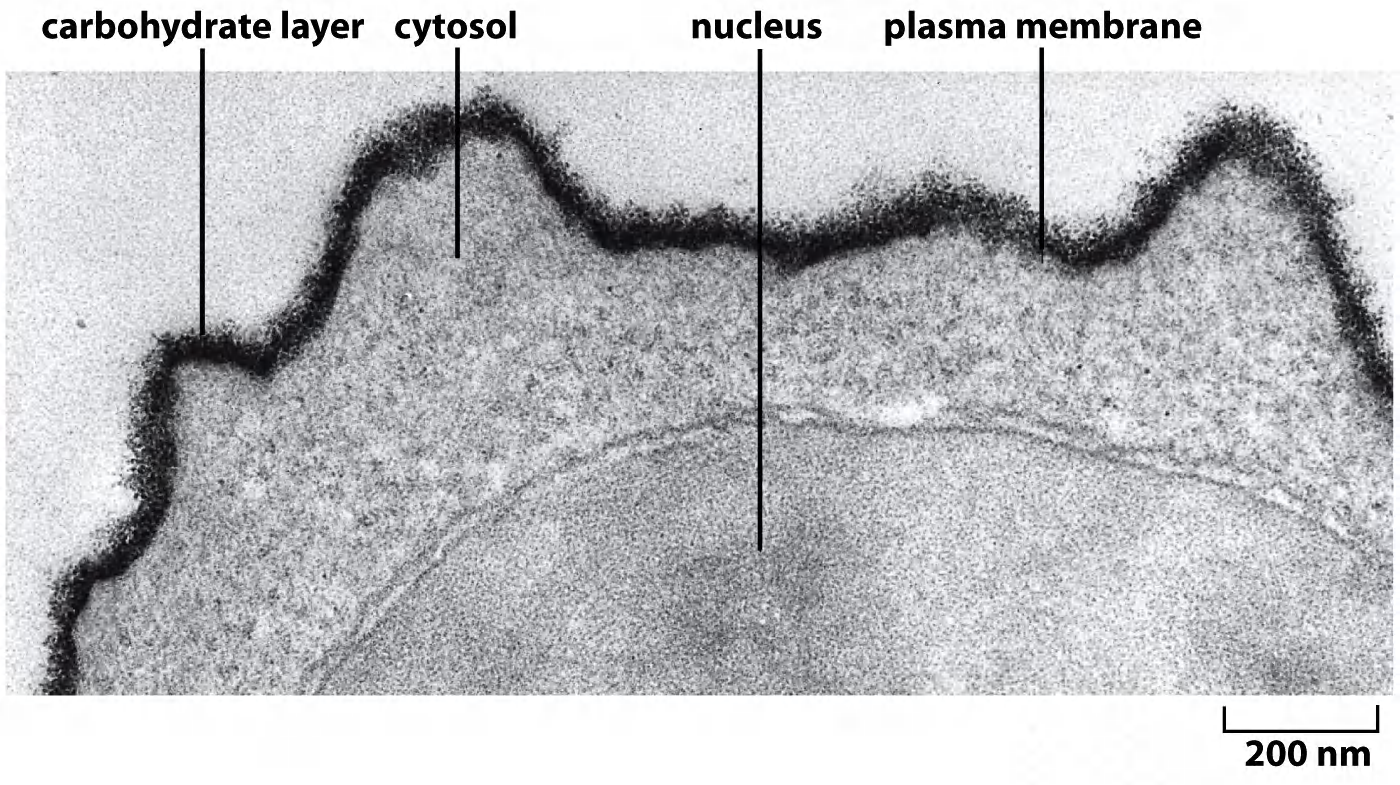
1. Carbohydrate layer visualized by ruthenium red stain (钌红染色)
Carbohydrate layer (Glycocalyx), also known as the pericellular matrix, is made of Glycoproteins and Glycolipids, surrounds the cell membranes of some bacteria, epithelia, and other cells, discovered by Martinez and Palomo 1970.
2. Lectins
lectins helps the neutrophil to migrate
二、Methods in membrane study
How to study membrane proteins?
Solubilization
Characterization: e. g. structural analysis
1. Detergents
In general, only agents that disrupt hydrophobic associations and destroy the lipid bilayer can solubilize membrane proteins
detergents, which are small amphiphilic molecules of variable structure
- Detergents are much more soluble in water than lipids. Their polar (hydrophilic) ends can be either charged (ionic), as in sodium dodecyl sulfate (SDS), or uncharged (nonionic), as in octylglucoside and Triton
when their concentration is increased above a threshold, called the critical micelle concentration (CMC), they aggregate to form micelles
- they also depend on the temperature, pH, and salt concentration.
Solubilization of membrane proteins by SDS
Strong ionic detergents, such as SDS, can solubilize even the most hydrophobic membrane proteins
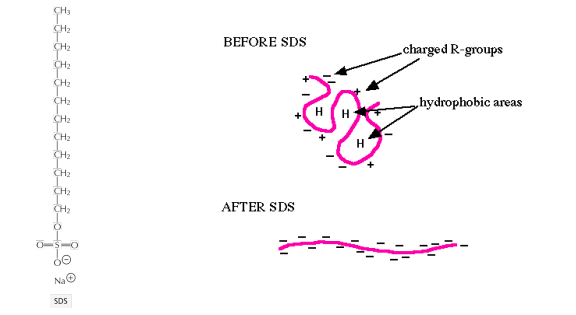
SDS fully denatures proteins
SDS covers protein molecules with negative charges
Charging the protein is essential for electrophoretic separation via SDS-polyacrylamide gel electrophoresis (PAGE)
Isolation of membrane proteins by non-ionic detergents
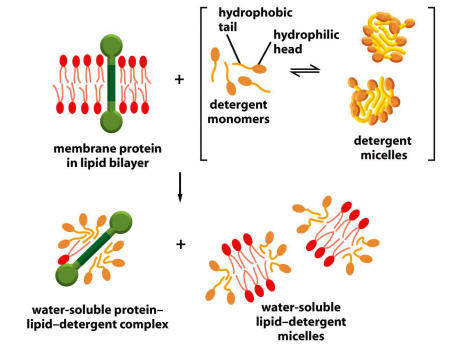
Non-ionic detergents only solubilize membrane components
- Remember the differences between ionic and nonionic detergents!
Functional reconstitution (重建) of proteins with the membrane
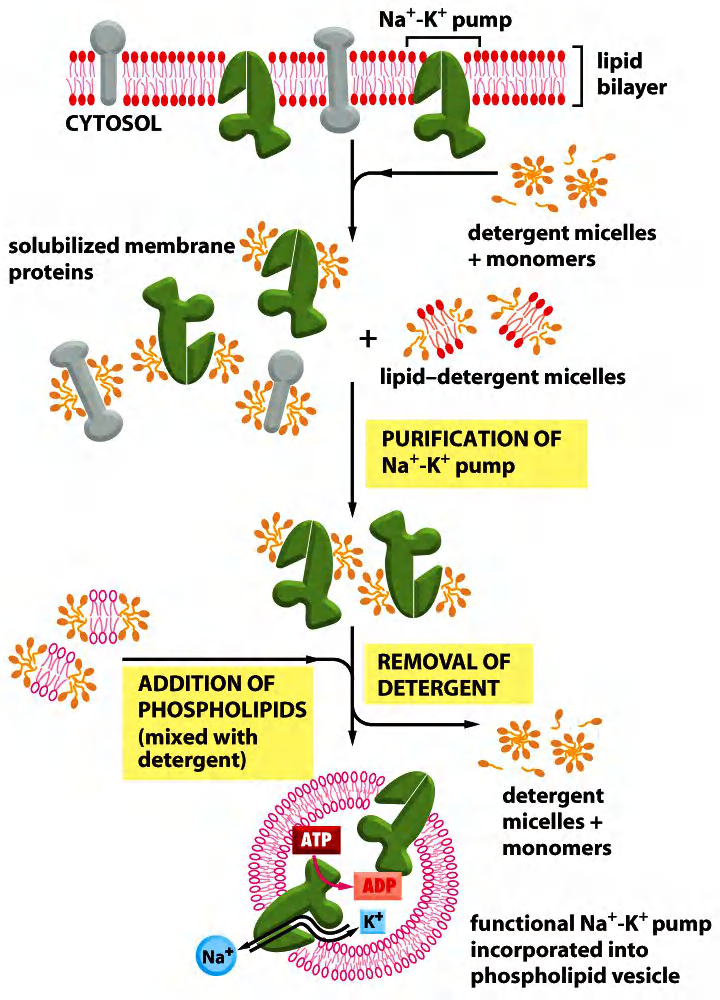
- First, the membrane lipids are substituted by the detergent molecules.
- Now, the detergent stabilizes protein structure (hydrophobic part).
- Next, the detergent is substituted by the added lipids.
- The protein is then in a liposome and can be further analyzed.
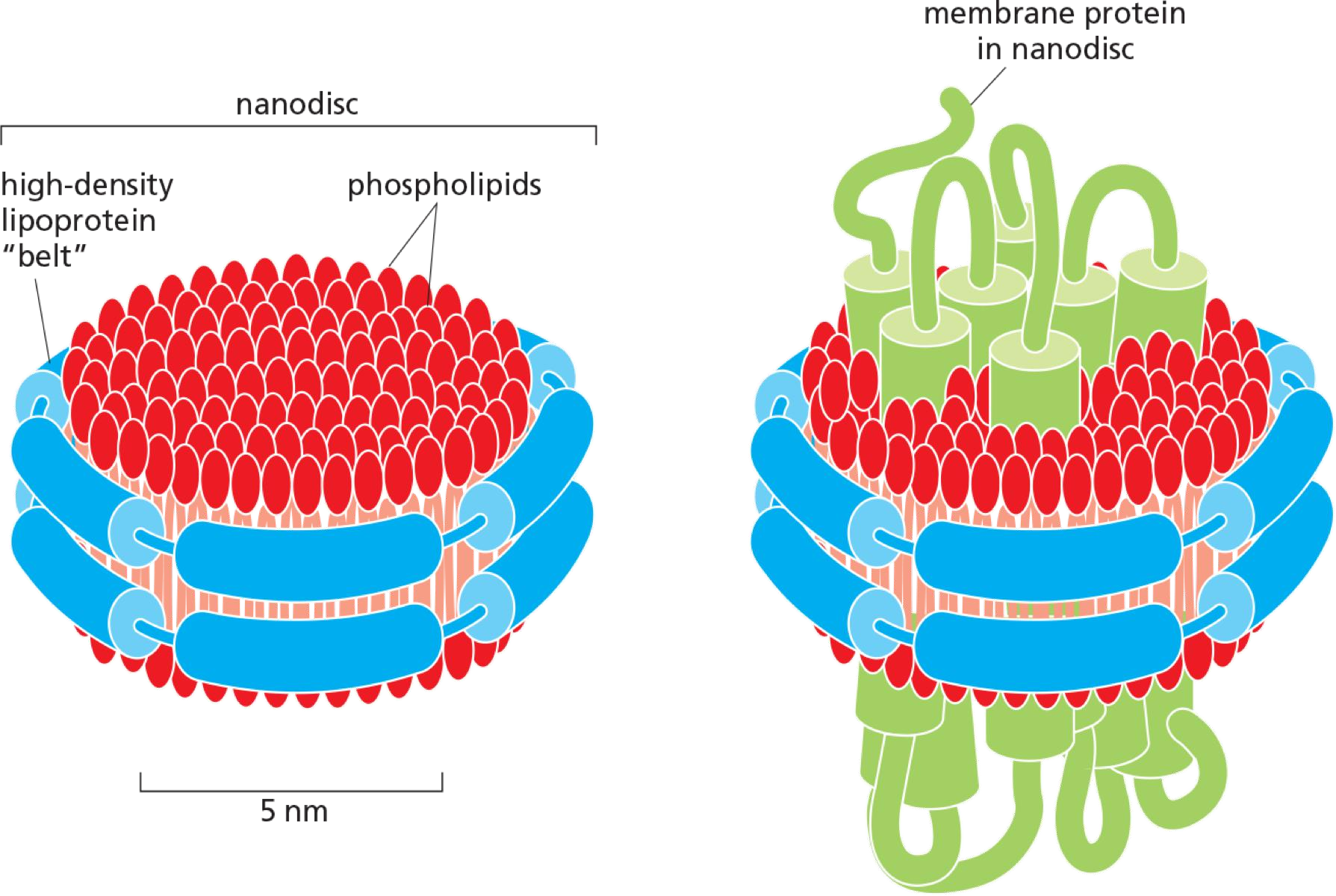
Nanodiscs: a small patch of lipid bilayer surrounded by a belt of highdensity lipoprotein (HDL,高密度脂蛋白), which shield the hydrophobic edges of the bilayer patch to render the assembly watersoluble.
A multi-pass membrane protein becomes embedded in the nanodisc.
The proteins can then be analyzed by single particle electron microscopy techniques to determine their structure.
Some Methods in Membrane Research
Cell-fusion
FRAP (fluorescence recovery after bleaching)
Functional reconstitution of membrane proteins with the membrane
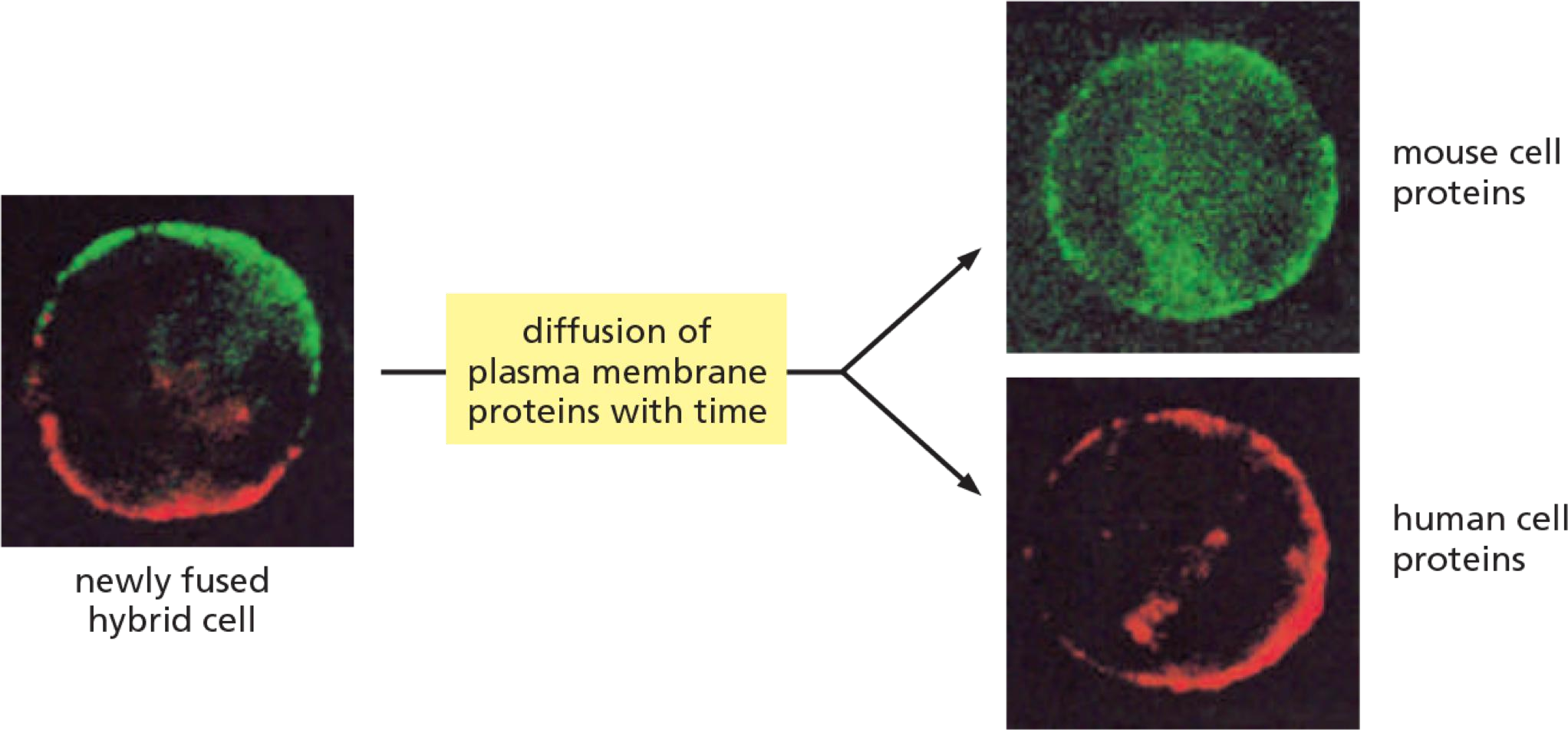
1. Experiment #1: How to proof movement of membrane proteins?
Example 1: Cell fusion experiment to detect lateral diffusion
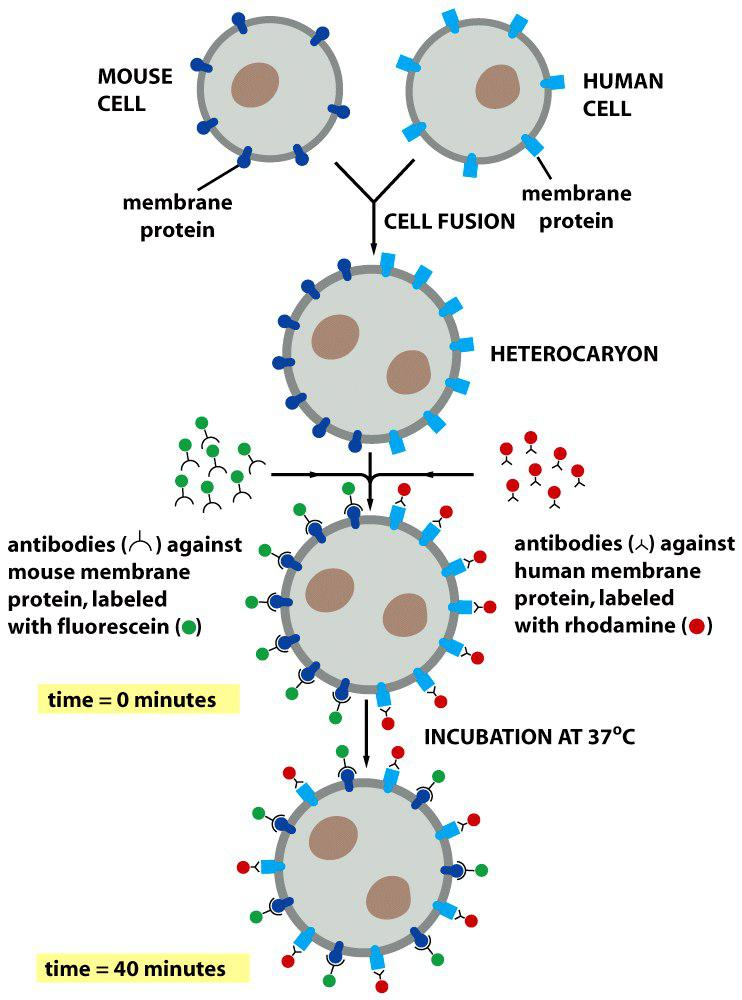
Analysis of lateral diffusion:
Immediately after fusion: Proteins are separated
After some time: proteins are mixed by diffusion
2. Experiment #2: How to proof movement of membrane proteins?
Example 2: Fluorescence recovery after photobleaching (FRAP) to detect diffusion
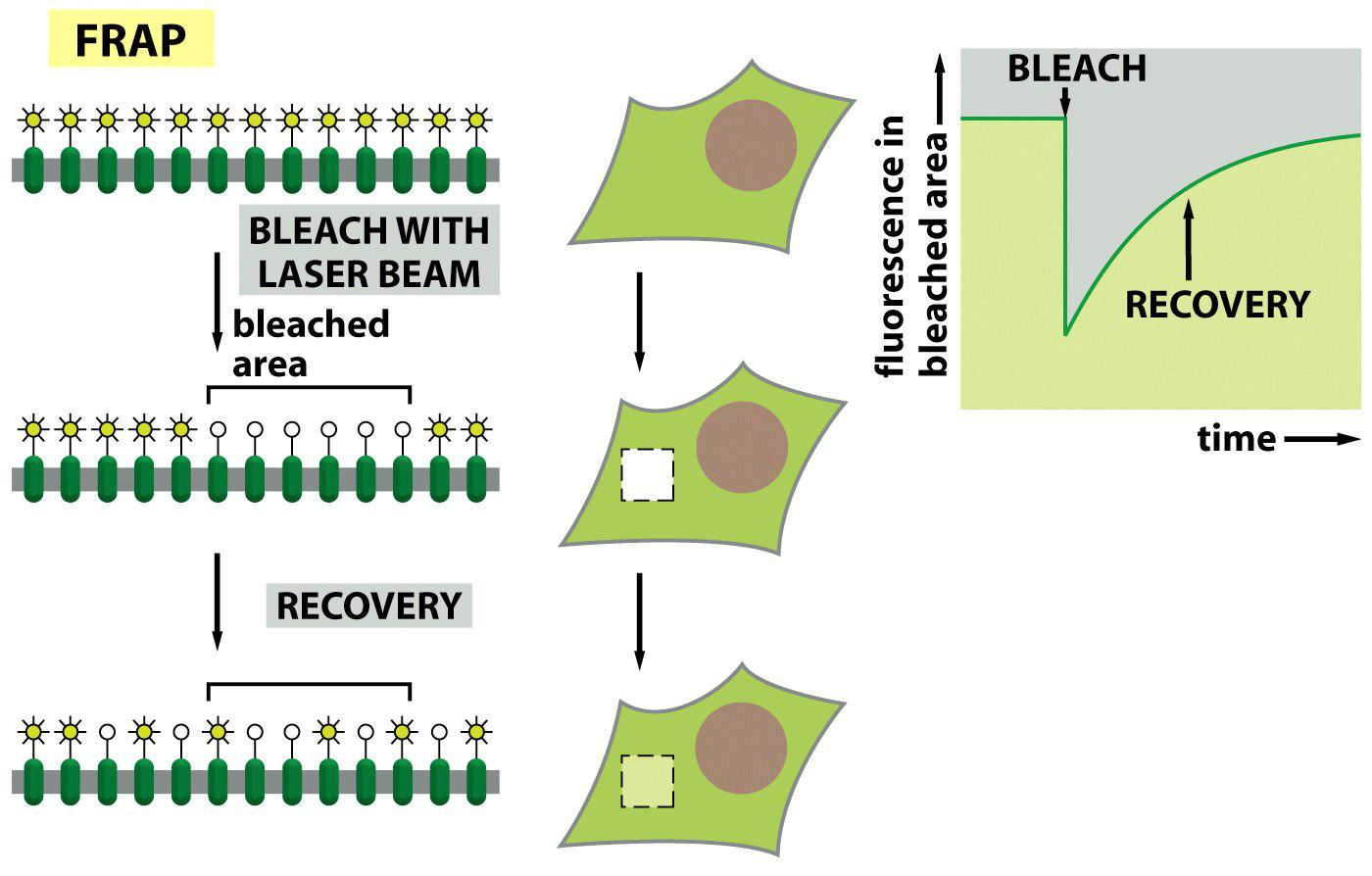
3. Experiment #3: How to proof movement of membrane proteins?
Example 3: Fluorescence loss in photobleaching (FLIP) to detect membrane protein diffusion
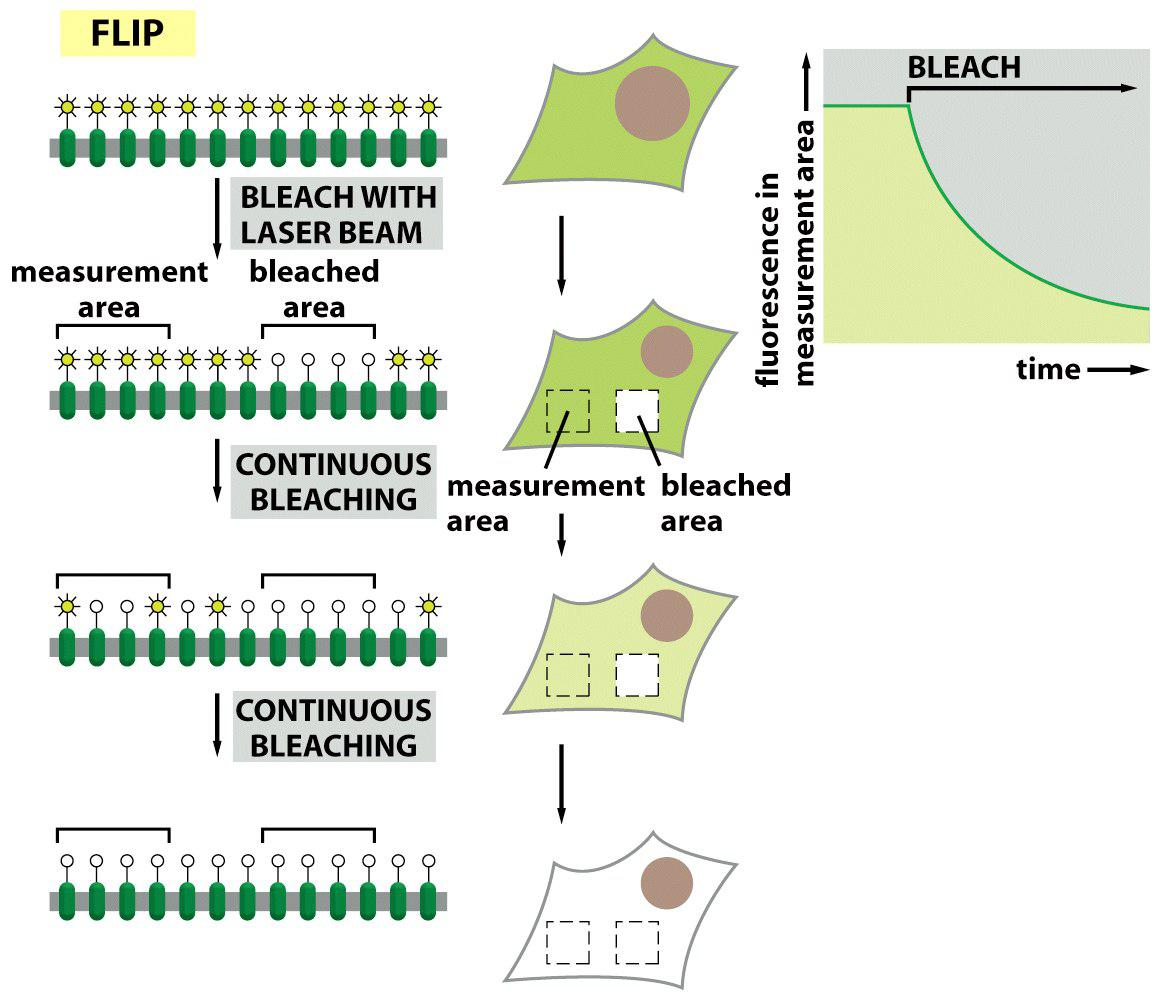
“Depletion of fluoresce” occurs if proteins from a “non-bleached” zone diffuse into the continuously ongoing bleaching zone because they are “bleached” there as well!
Do all membrane proteins freely move?
