L09 Structure of DNA and RNA
一、Nucleotide And Polynucleotide Chain Structure
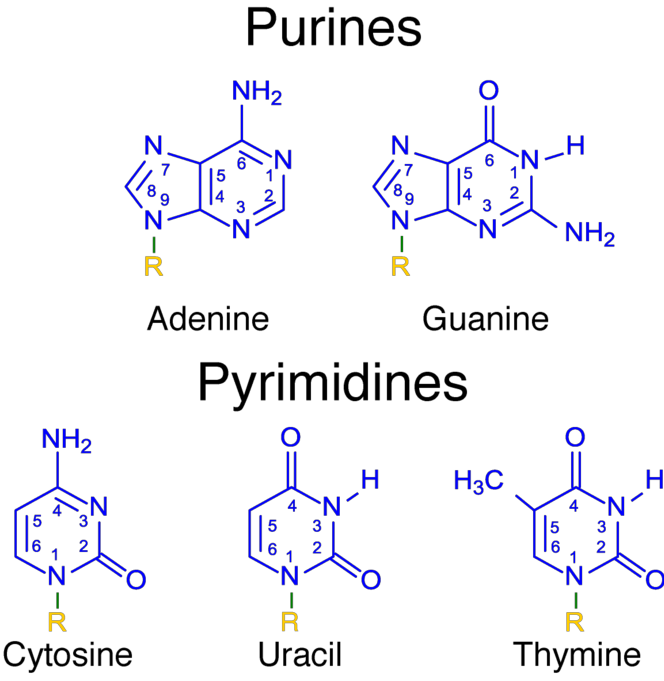
Nucleoside (核苷) And Nucleotide (核苷酸)
1. Nucleoside
Adenine + ribose = Adenosine
Adenine + deoxyribose = Deoxyadenosine
2. Nucleotide
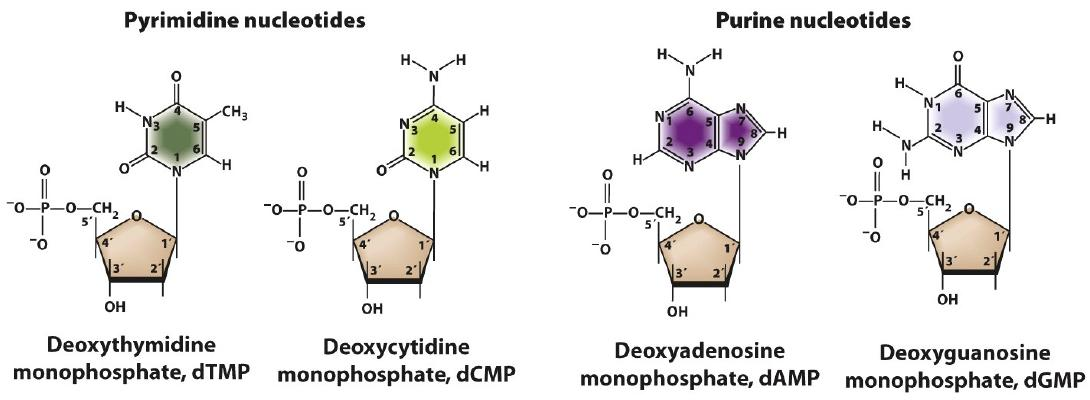
Base + sugar + phosphate(s) nucleotide
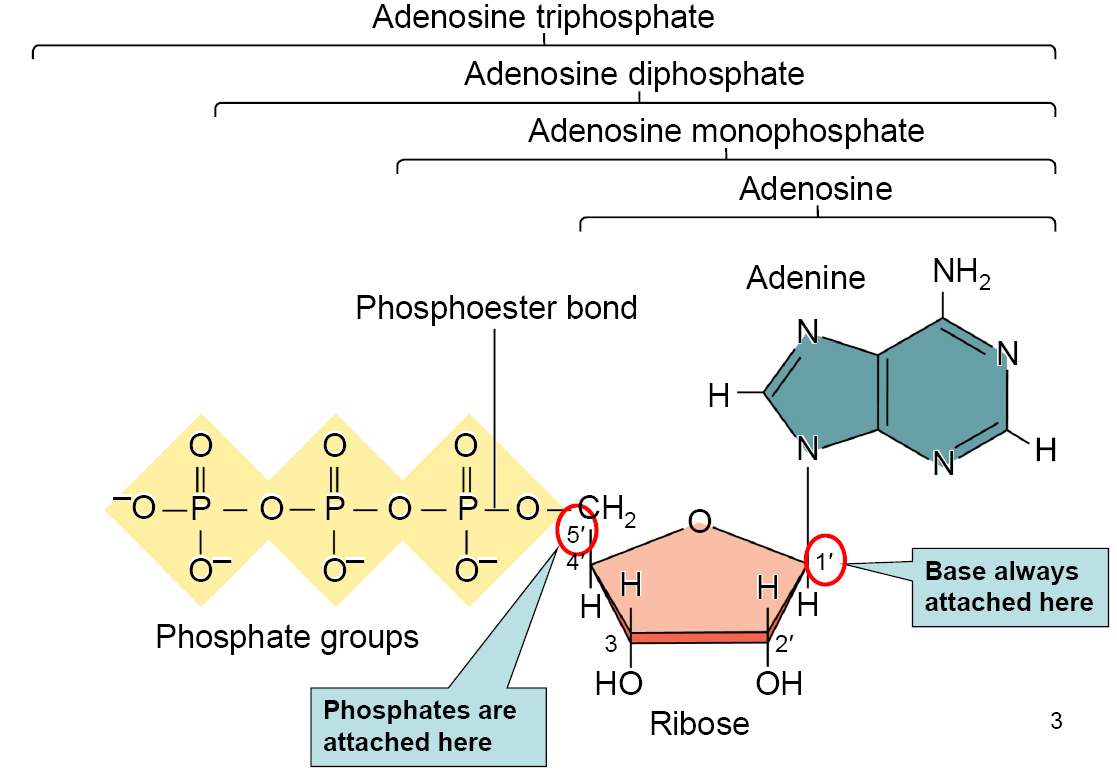
Adenosine monophosphate (AMP) = Nucleoside + $P$
Adenosine diphosphate (ADP) = Nucleoside + $PP_{i}$
Adenosine triphosphate (ATP)
DNA Polynucleotide Chain Structure
Nucleotides are covalently linked together by phosphodiester bonds (磷酸二脂键)
A phosphate connects the 5’ carbon of one nucleotide to the 3’ carbon of another
The Direction
The chain has 5’ to 3’ direction because the phosphodiester bonds has its direction
Backbone
The phosphates and sugar molecules form the backbone of the nucleic acid strand
The bases project from (从…伸出) the backbone.
1. Hydrogen-bonds
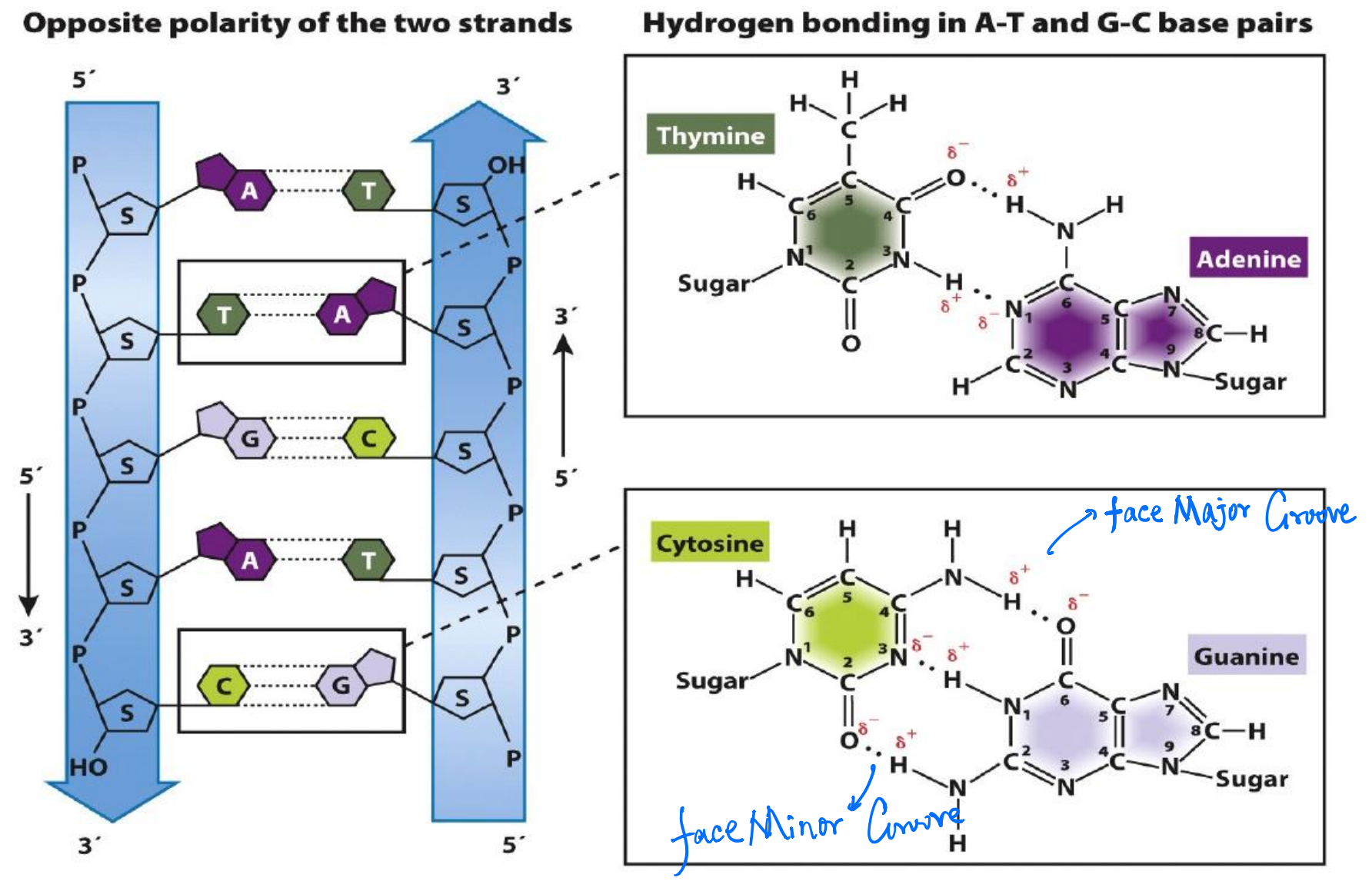
2. Double Helix
- Two strands are twisted together around a common axis
- There are 10 bases and 3.4 nm per complete turn of helix
- The two strands are antiparallel
- The helix is right-handed
- Hydrogen-bonding between complementary bases
Two grooves
There are two asymmetrical grooves on the outside of the helix
Major groove and Minor groove
- Certain proteins binds to these grooves, thus they interacts with a particular sequence of bases
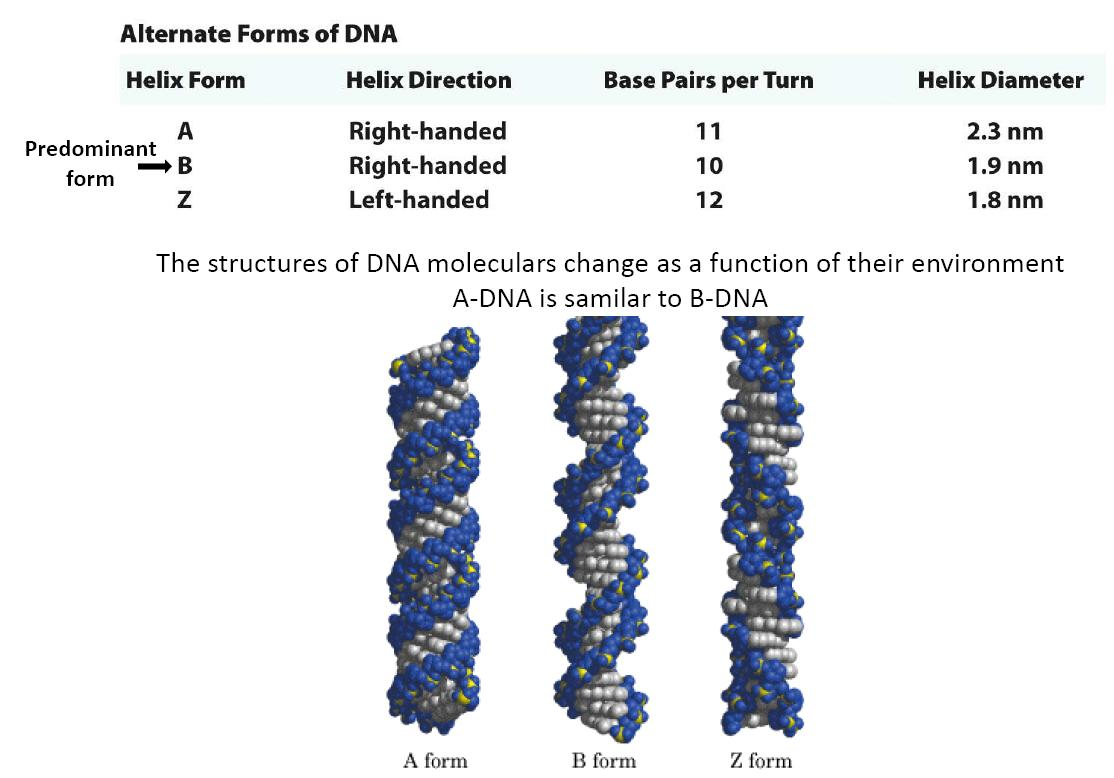
Forms of Double Helix
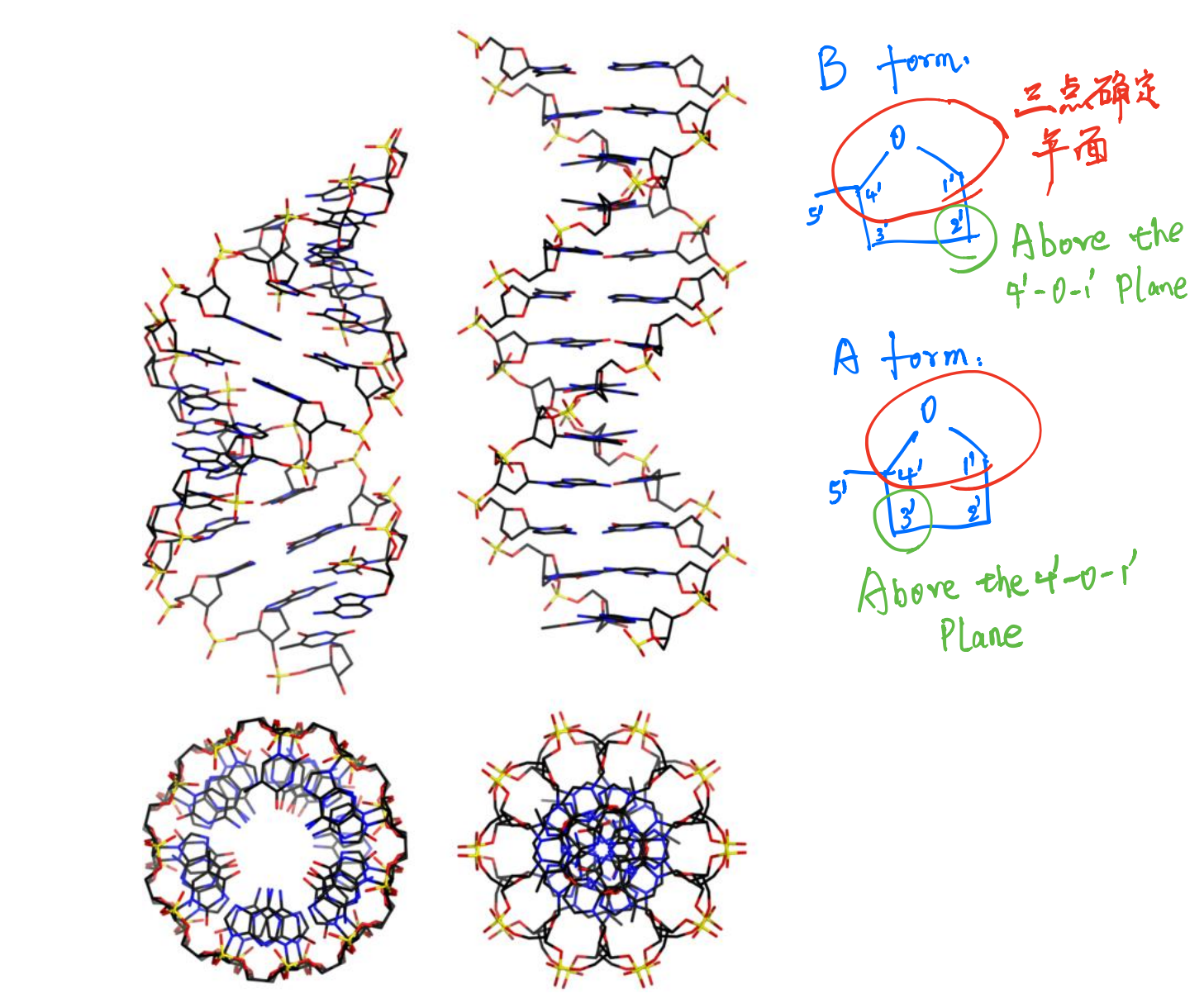
Sugar Pucker
3. Chromatin And Chromosome
Human genome (in diploid cells) = 6 x $10^{9}$ bp
6 x $10^{9}$ bp X 0.34 nm/bp = 2.04 x $10^{9}$ nm = 2 m/cell, but diameter of nucleus = 5-10 μm
So DNA must be packaged to protect it, but must still be accessible to allow gene expression and cellular response, the solution is cheomosome
Chromosome
Chromosome is the complex of single DNA molecular and associated proteins
- Evident (明显的) during metaphase of Mitosis and Meiosis
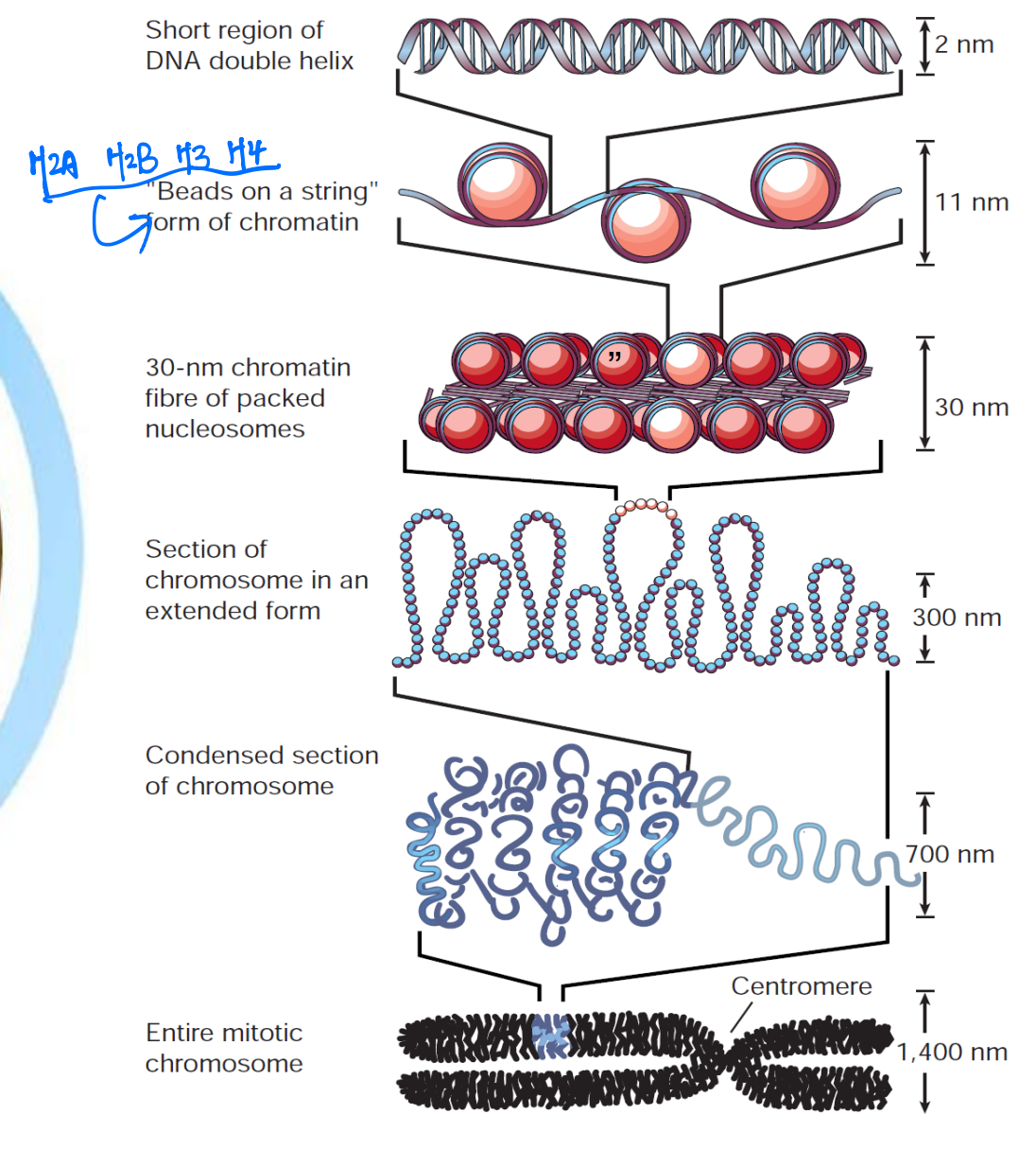
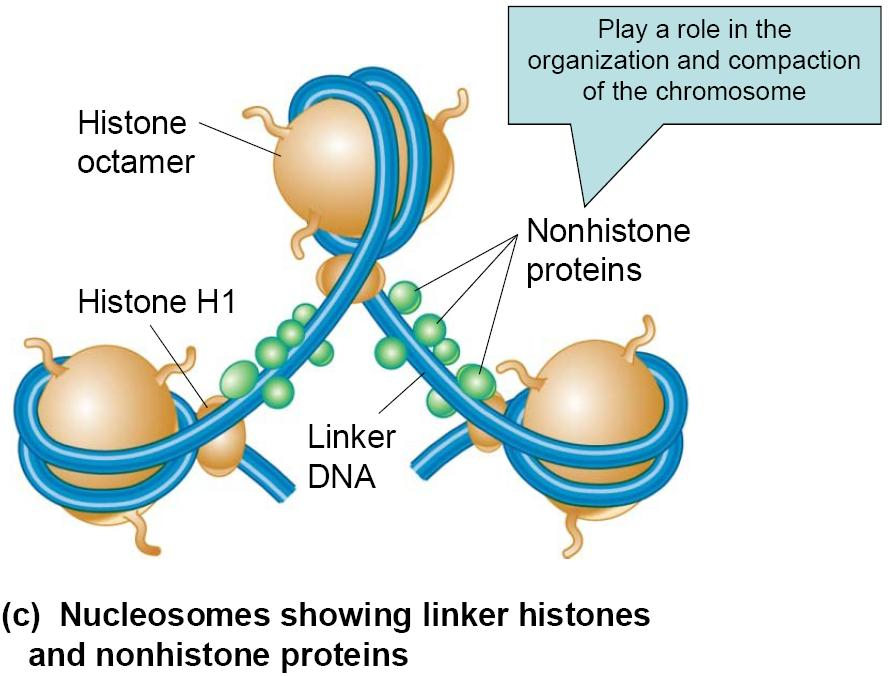
The repeating structural unit within eukaryotic chromatin is the nucleosome
It is composed of a double-stranded segment of DNA wrapped around an octamer of histone proteins
A histone octamer is composed of two copies each of four different histone proteins
146 bp of DNA make negative superhelical (左旋) turns around the octamer
DNA is 1.75 turns per histone
Enzymatic digests
Particular enzymes can cut double stranded DNA between basepairs
Most of the enzymes have specific recognition sites
(1) MNase digests
Particular enzymes can cut double stranded DNA between
- Micrococcal nuclease (MNase) cleaves everything it can (no ⾮限制性 basepairs specific seq.)
This micrococcal nuclease is a stable enzyme derived from Staphylococcus aureus. Micrococcal nuclease exhibits exo- and endo-5’-phosphodiesterase activities against DNA and RNA. This enzyme digests double-stranded, single-stranded, circular and linear nucleic acids.
Micrococcal nuclease cuts accessible regions of DNA
DNA protected by nucleosome is not cut
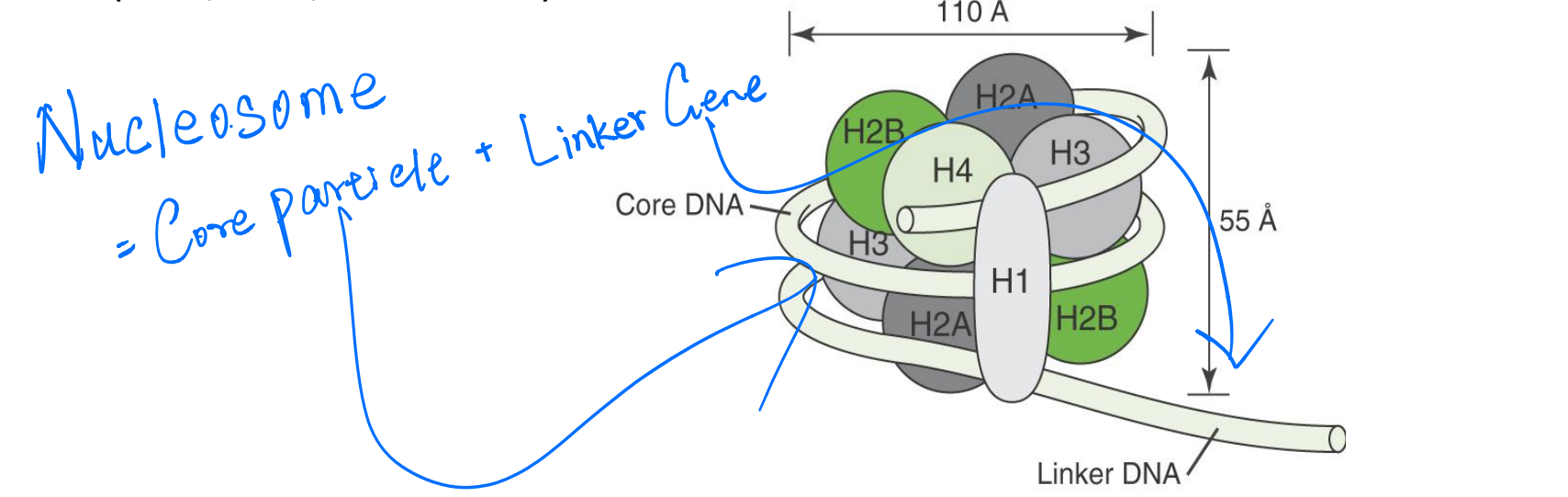
Structure of individual nucleosome
Nucleosome is the basic structural subunit of chromatin, consisting of ~200 bp of DNA, an octamer of histones (H2A, H2B, H3 and H4), and H1.
Core DNA is the 146 bp of DNA contained on a core particle.
Linker DNA is all DNA contained on a nucleosome in excess of the 146 bp core DNA.
Nucleosome core particle (NCP) is the core including 146 bp DNA and histone octamer (H2A, H2B, H3 and H4)
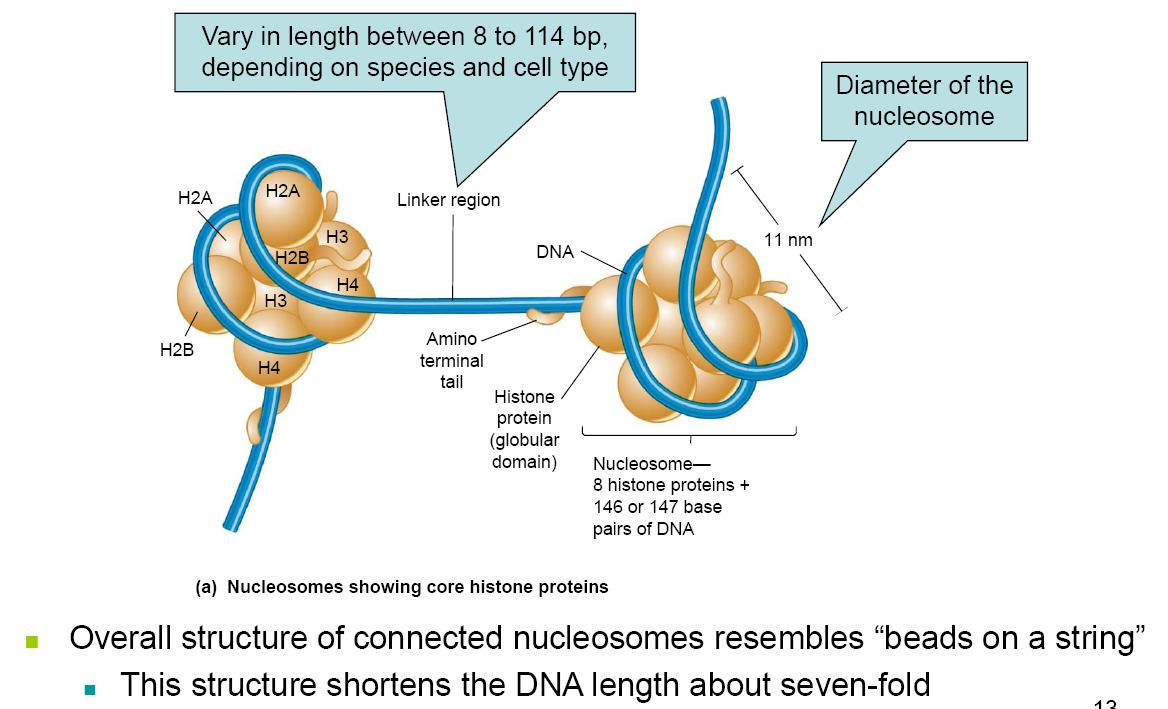
Repeating structure contains 5 different proteins:
- H1, H2A, H2B, H3, H4
- Core structure formed from: H2A, H2B, H3, H4 (Core histones)
- H1 is called the linker histones
- Binds to DNA in the linker region
- Less tightly bound to DNA than core histones
- May help compact adjacent nucleosomes
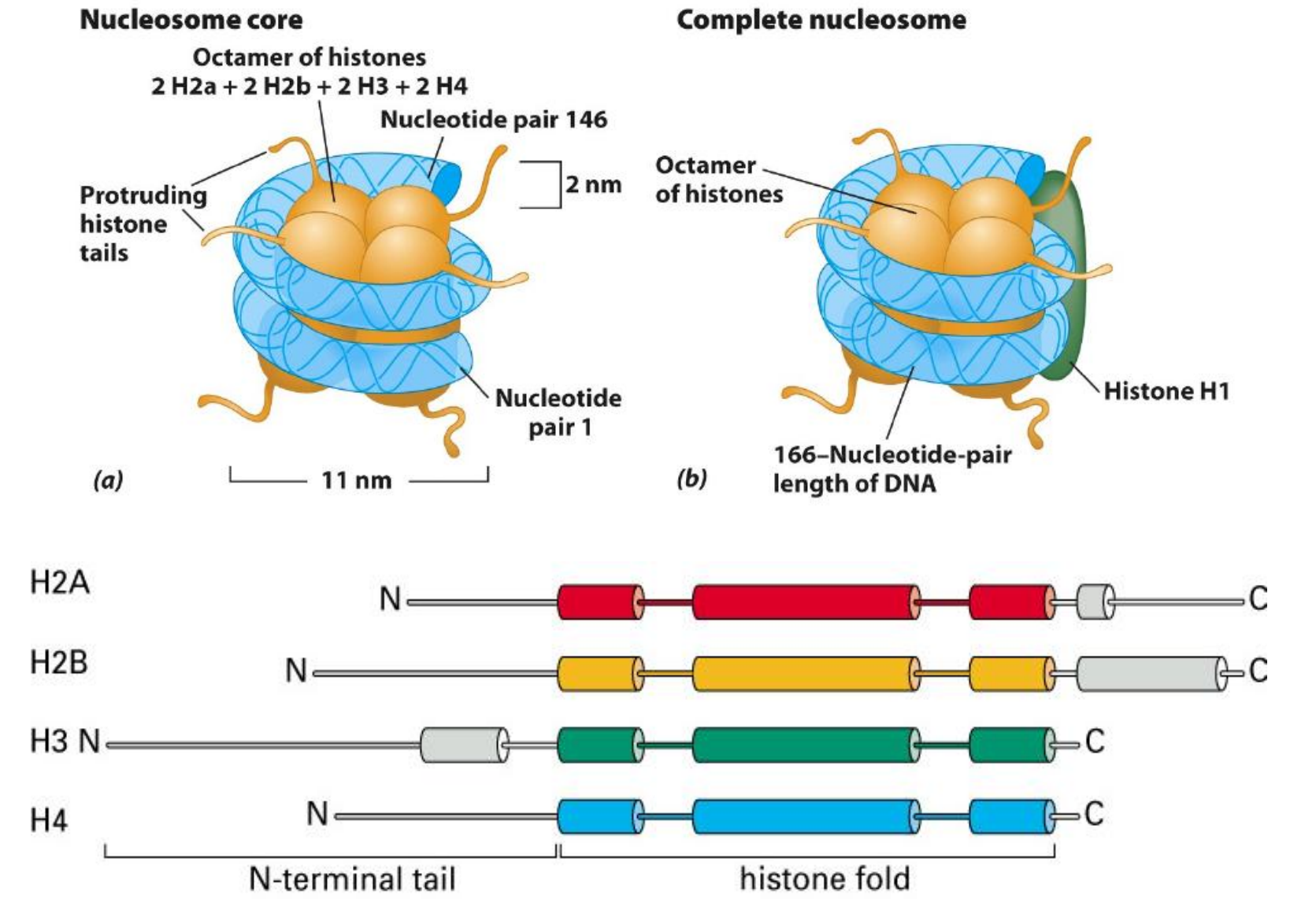
(1) The Connection Between Histone And DNA Molecular
The histone proteins are basic
- They contain positively-charged amino acids
- Lysine and Arginine
- H2A and H2B are more negatively-charged, Virus can use this site.
- They binds to the phosphate group (Negatively-charged) on the DNA molecular
Histone has a globular domain and a flexible, charged amino acid terminus or ‘tail’
Epigenetic make modifications on the ‘tail’.
(2) Nonhistone proteins
RNA Polynucleotide Chain Structure
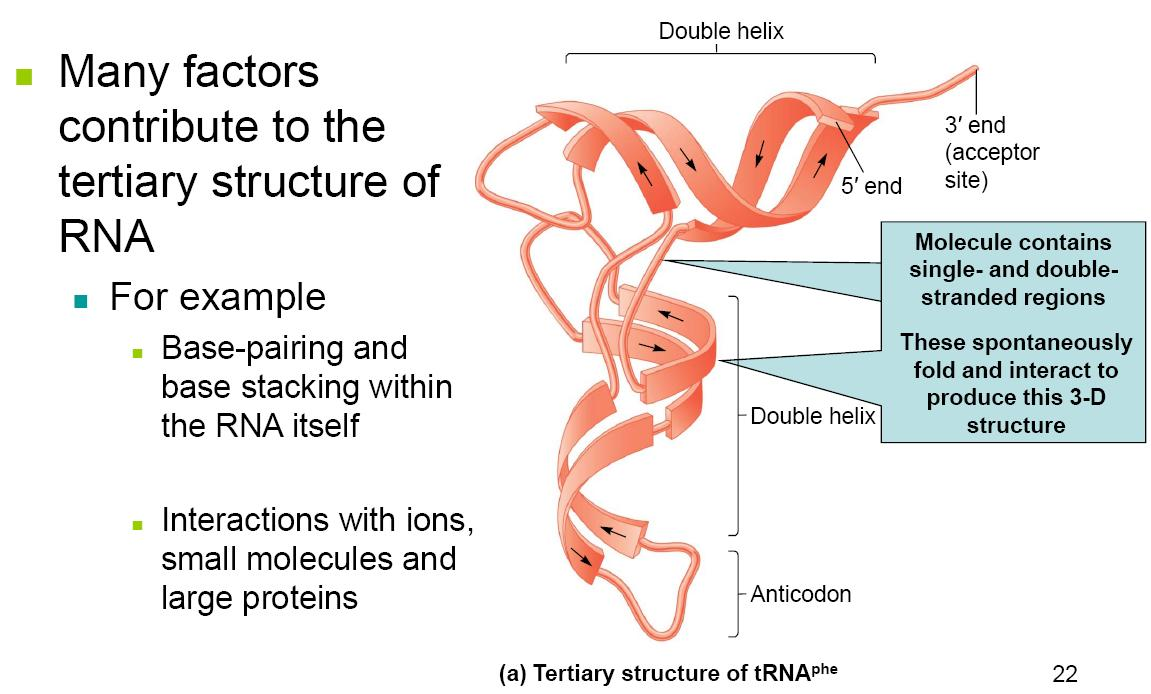
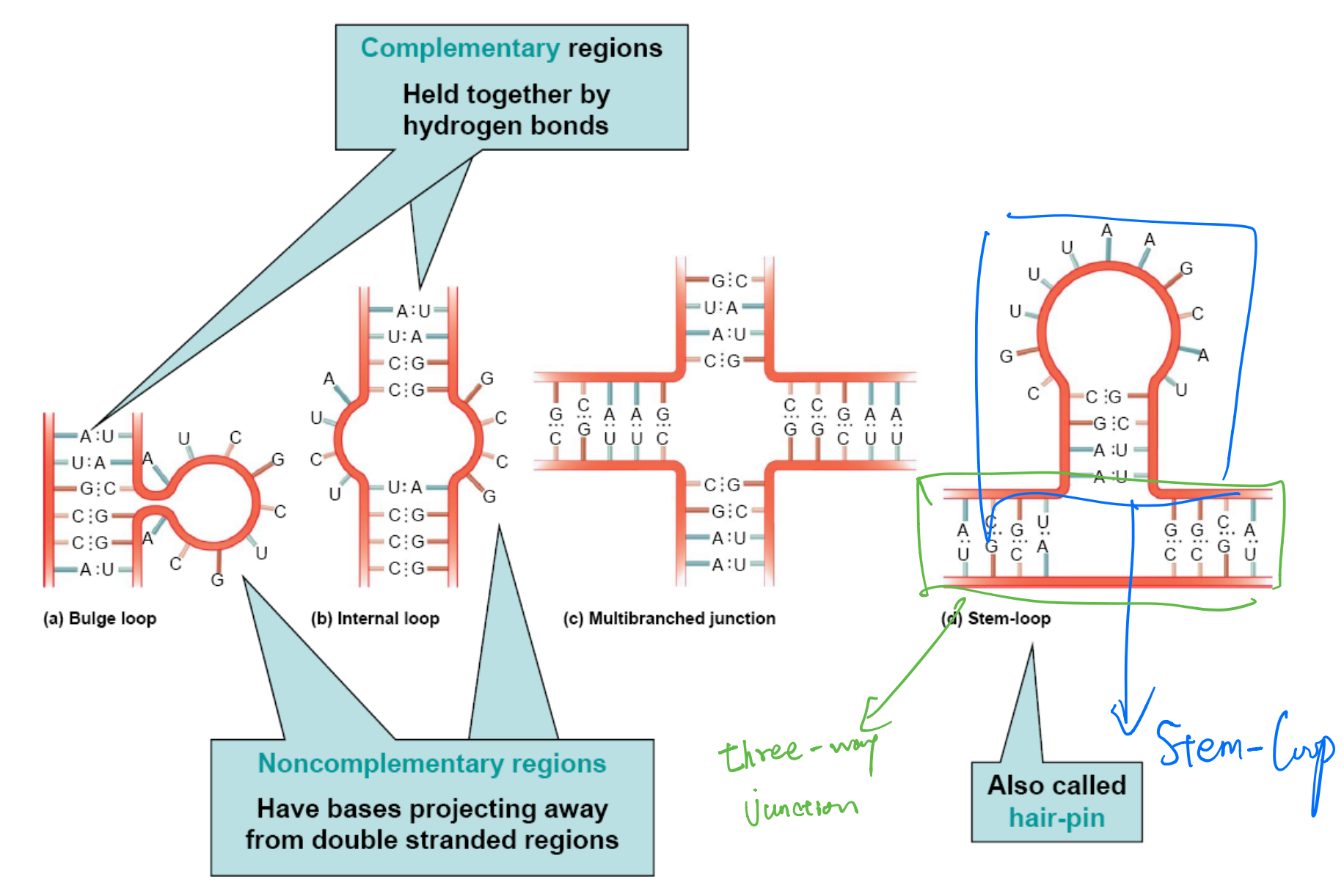
Although usually single-stranded, RNA molecules can form short double-stranded regions
- This secondary structure is due to complementary base-pairing
- A to U and C to G
- This allows short regions to form a double helix
RNA helices are typically right-handed, 11-12 bases per turn in the A form
1. Secondary Structure
2. Tertiary Structure