L06 peptide and protein structure
Peptides and Peptide bond
Peptides
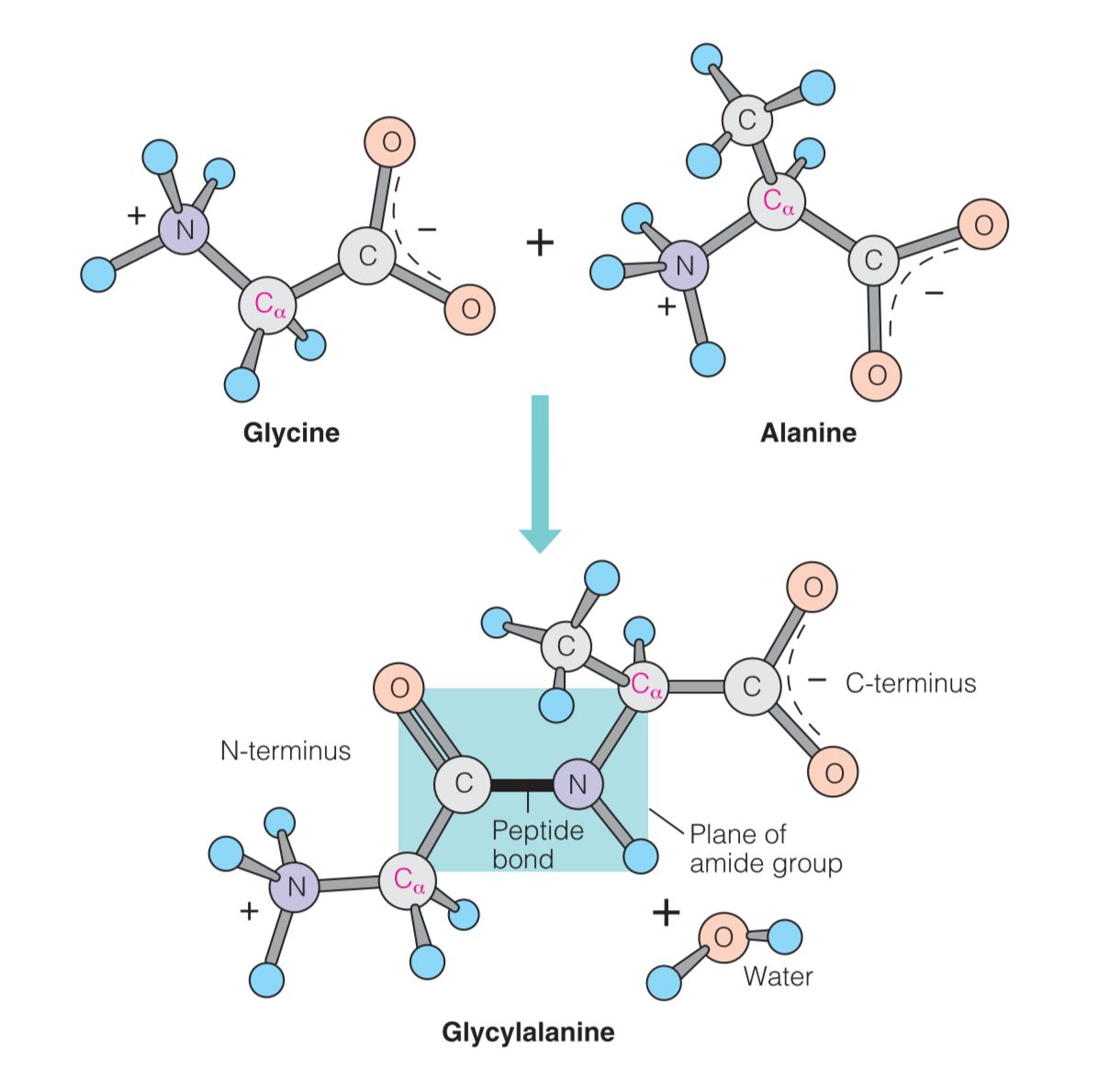
The portion of each amino acid remaining in the chain is called an amino acid residue(氨基酸残基)
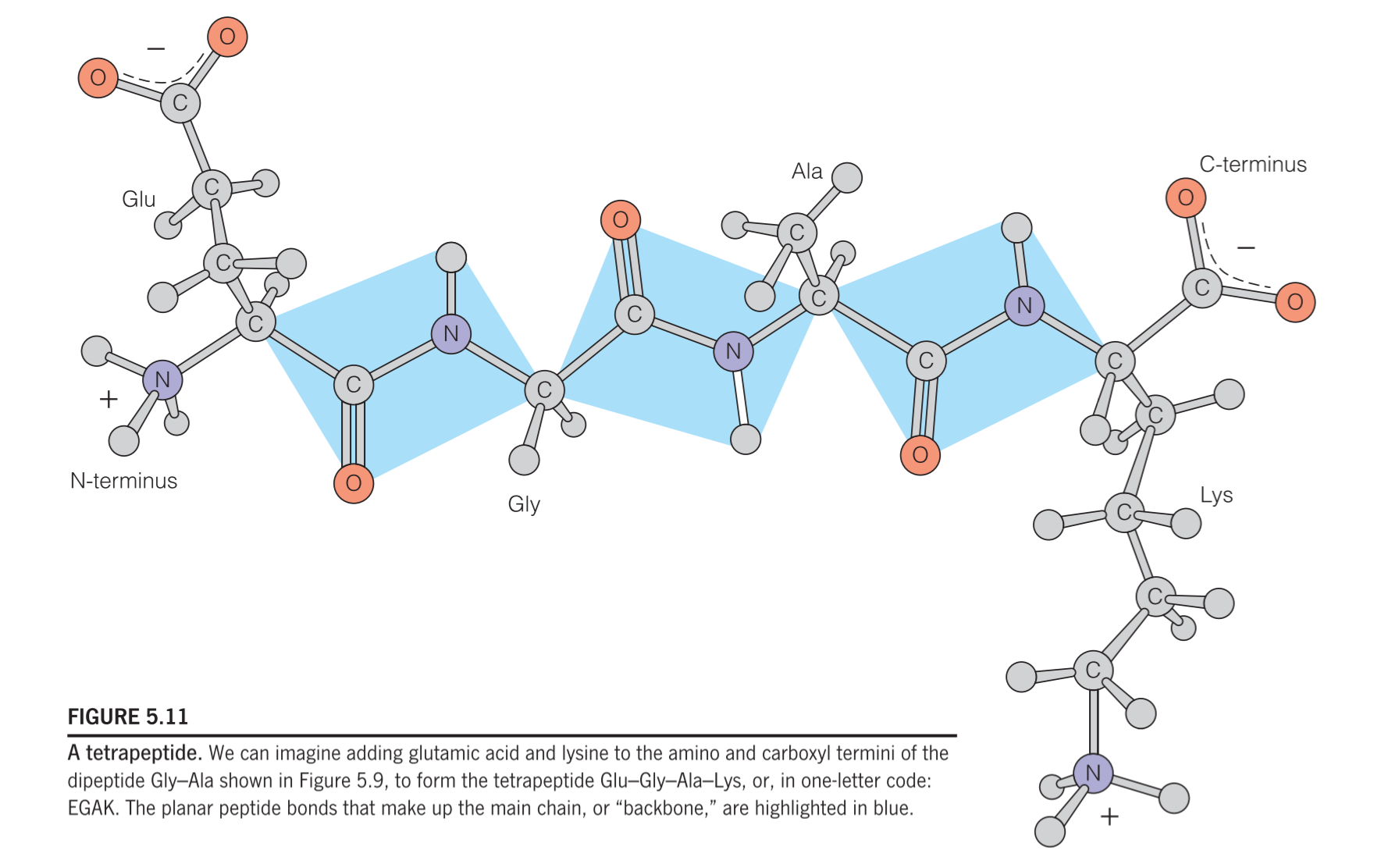
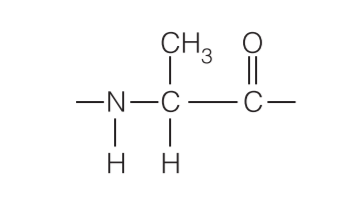
When specifying an amino acid residue in a peptide, the suffix –yl may be used to replace –ine or –ate in the name of the amino acid (e.g., glycyl for glycine, aspartyl for aspartate; tryptophanyl and cysteinyl are exceptions to this general rule). Thus, the alanyl residue in the tetrapeptide in Figure 5.11 is:
1. Residue number influence the name
- Chains containing only a few amino acid residues (like a tetrapeptide) are collectively referred to as oligopeptides.
- If the chain is longer (>~15–20 residues), it is called a polypeptide.
- Polypeptides greater than 50 residues are generally referred to as proteins.
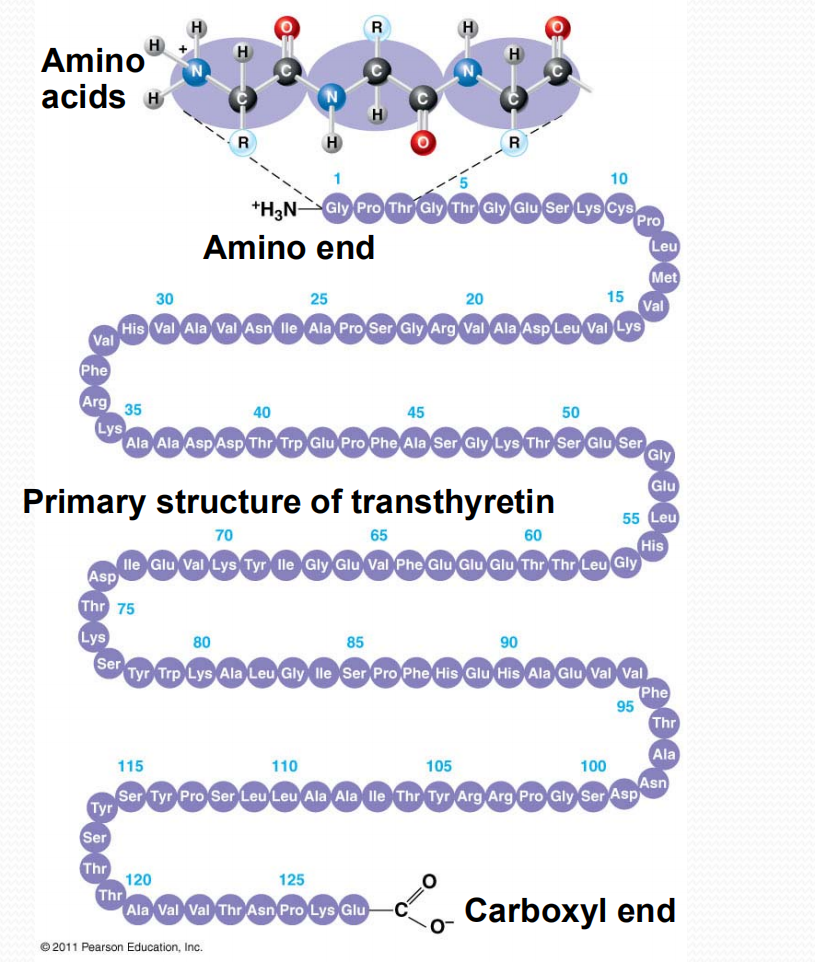
As shown in Figure 5.11, most oligopeptides and polypeptides retain an unreacted amino group at one end (called the amino terminus or N-terminus) and an unreacted carboxylic acid group at the other end (the carboxyl terminus or C-terminus).
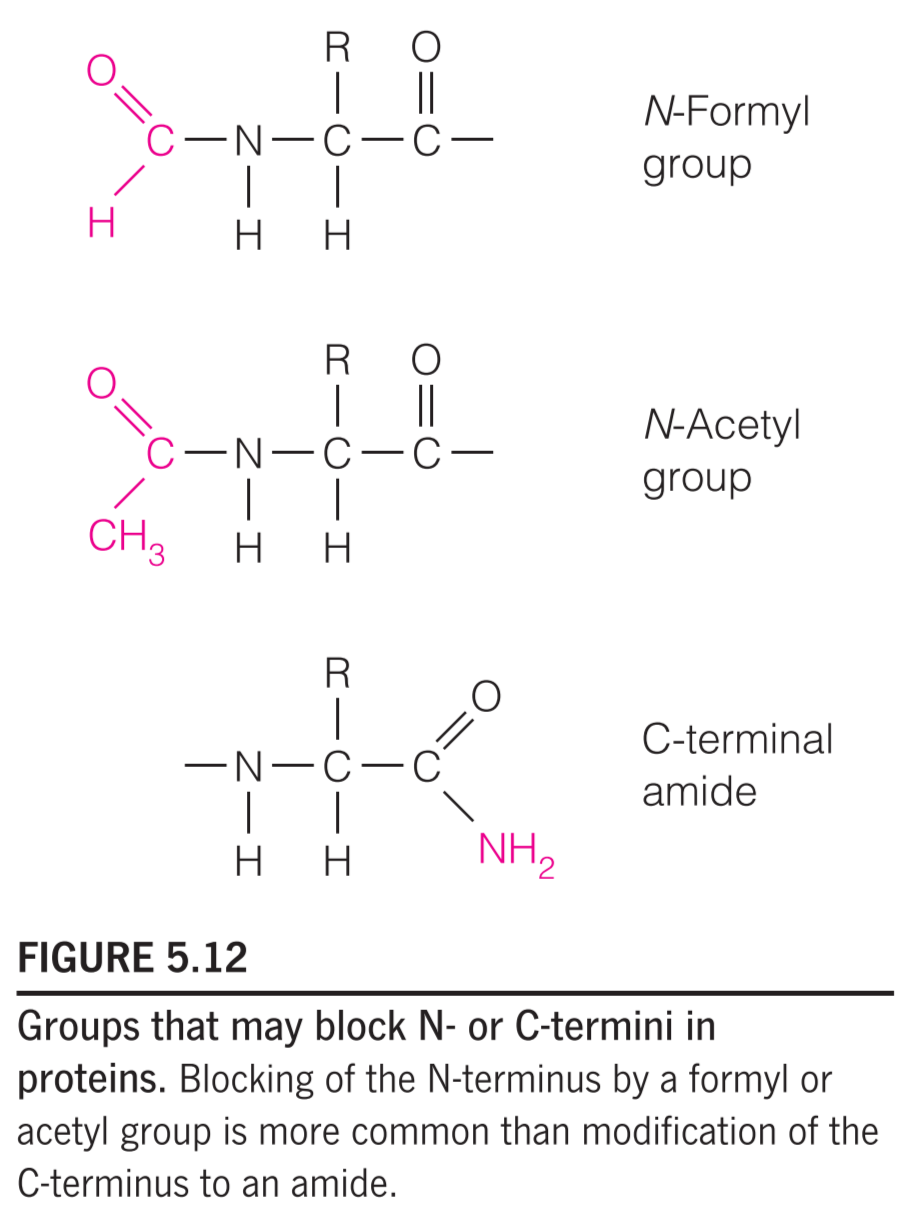
In addition, many proteins have N-termini blocked by N-formyl or N-acetyl groups, and a few have C-terminal carboxylates that have been modified to amides (Figure 5.12).
Polypeptides as Polyampholytes(两性电解质)
polypeptides usually contain some amino acids that have ionizable groups on their side chains, these groups have a wide range of pKa, but are all weakly acidic or weakly basic groups.
amino acid side chains display a range of pKa values in different proteins due to differences in the local electrostatic environment around a given amino acid.
For example, an aspartic acid side chain near another negatively charged group (i.e., Glu or another Asp) will likely have a higher value than an aspartic acid near a positively charged group (i.e., Lys, His, or Arg).
1. Polypeptides as polyampholytes – isoelectric point
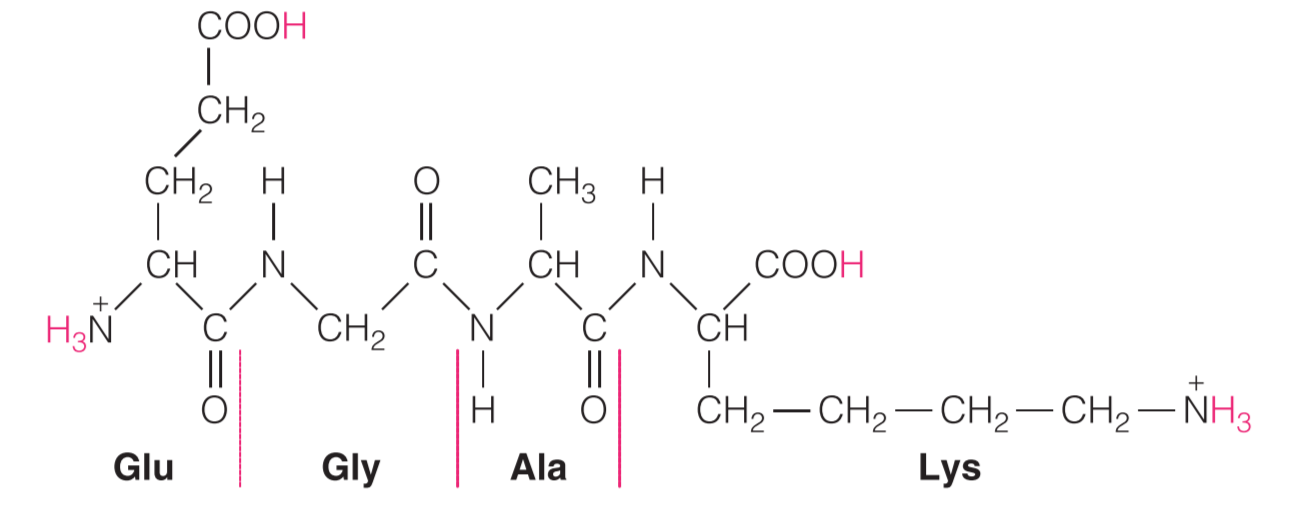
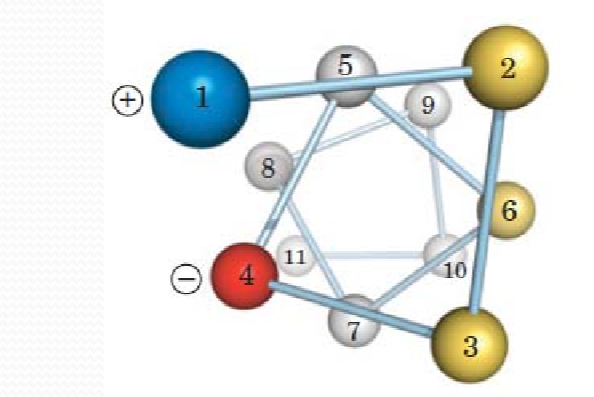
Example;the titration of an oligopeptide or a polypeptide is exemplified by the tetrapeptide (Glu-Gly-Ala-Lys),如下图
在初始条件下(PH=0),所有的可电离残基都处于它们的protonated forms,因此带电量为+2
As more base is added, the molecule becomes increasingly negatively charged, ultimately reaching a net charge of at very high pH.
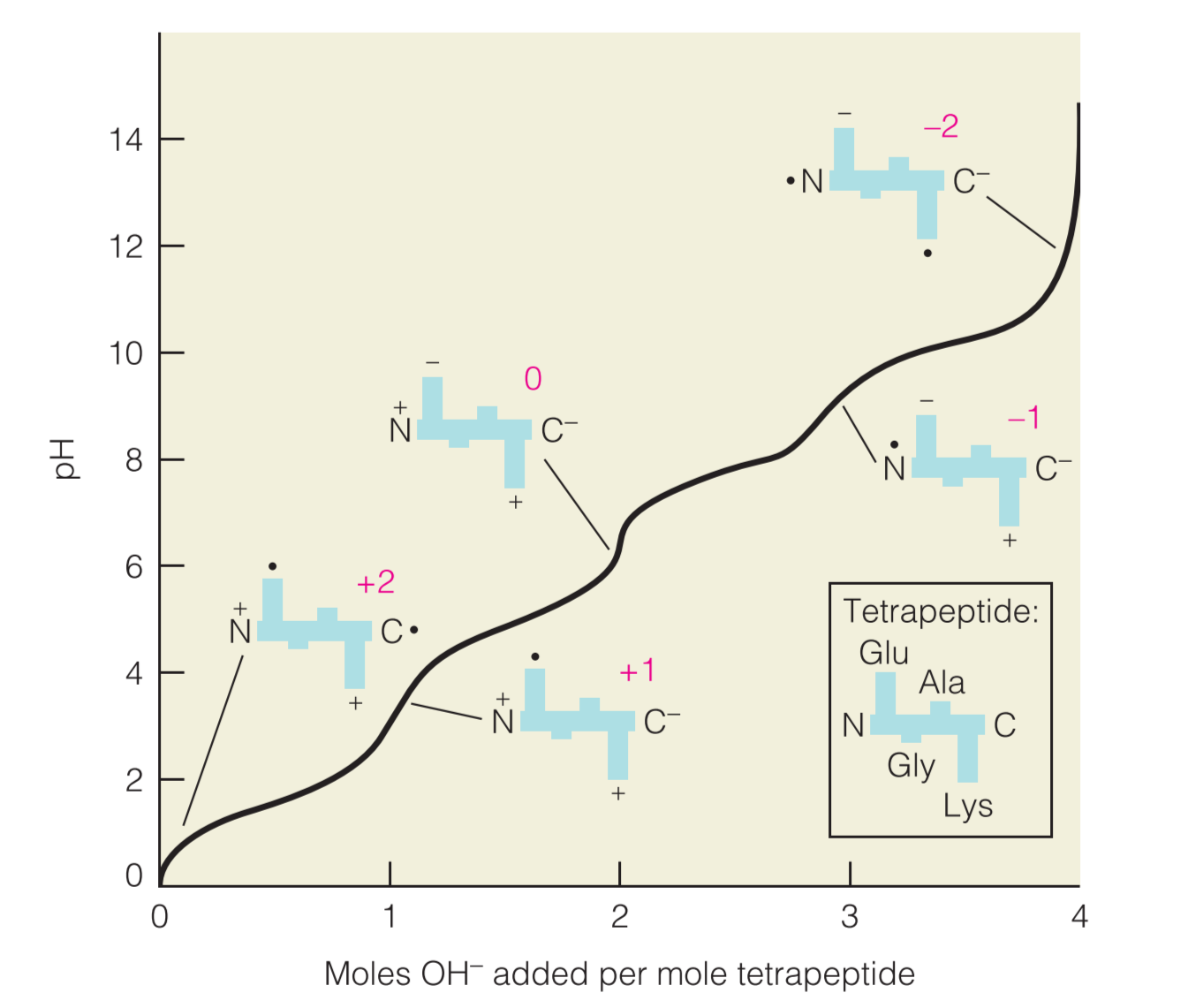
改变PH环境对于蛋白质具有重要影响:
- small shift in pH can alter the constellation of charges displayed on the surface, or in the active site, of a protein and will thereby significantly affect its stability and/or functional properties.
- the solubility of many proteins is minimal at the isoelectric point because the molecules no longer repel one another when their net charge is zero
Peptide Bond
the blue-shaded portion contains the peptide bond. This substituted amide bond, which is found between every pair of residues in a protein, has properties that are important for defining the structures of proteins.
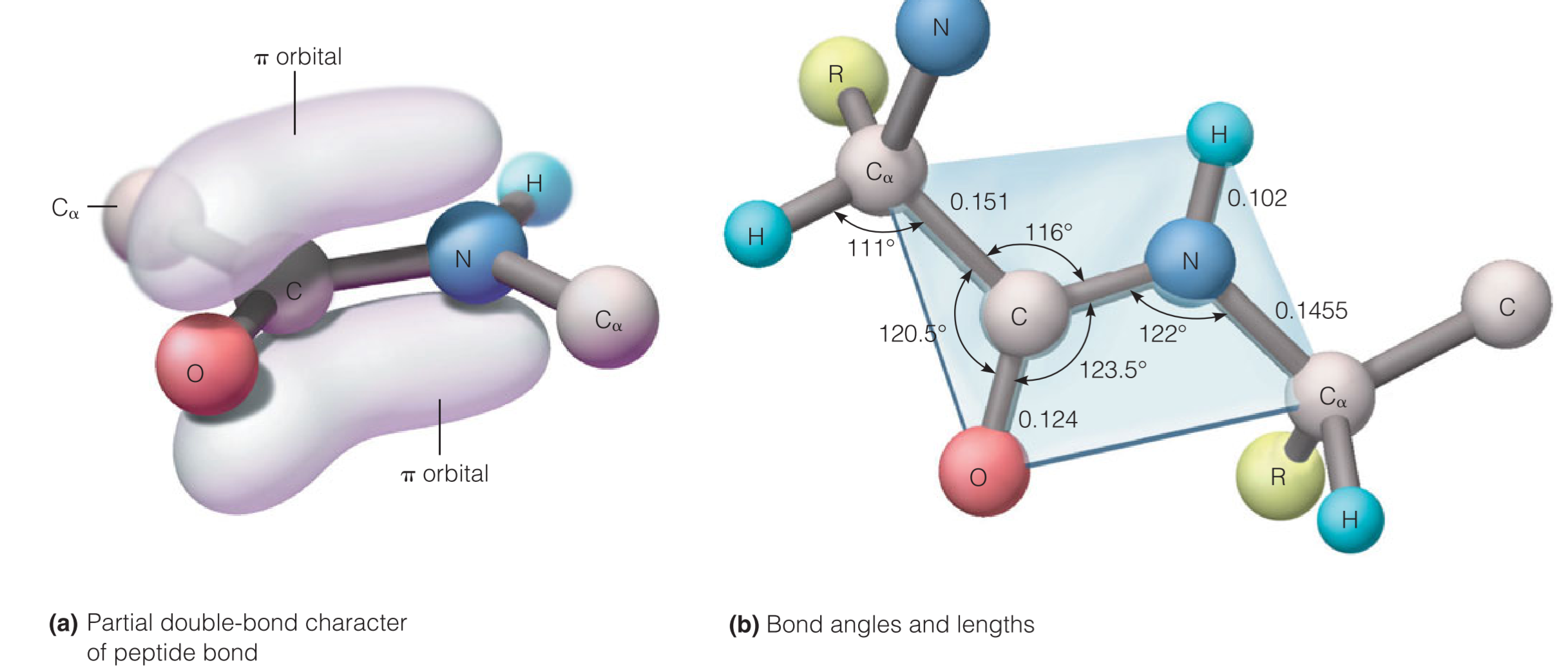
The amide carbonyl and amide bonds are nearly parallel
The six atoms shown in the blue rectangle in usually coplanar.
There is little twisting possible around the peptide bond because the bond has a substantial fraction(很大的一部分) of double-bond character.
肽键C–N之间的距离为1.33$\AA$
Amino acid residue
Include:
- Mainchain (peptide backbone)
- Sidechain
Directional linkage
Amino group $\rarr C_{\alpha}\rarr$Carbonyl group
Amino group is the N-terminus (amino terminus) and Carbonyl group is the C-terminus (carboxyl terminus)
Rotation
Rotation around peptide bond is restricted
Conformation(构造)of peptide bond
(1) Backbone
-[N-$C_\alpha$-C]-
(2) Dihedral angles 二面角
Rotation about both the N-$C_\alpha$ ($\phi$) and $C_{\alpha}-C$($\psi$) bonds is possible
1. resonance hybrid
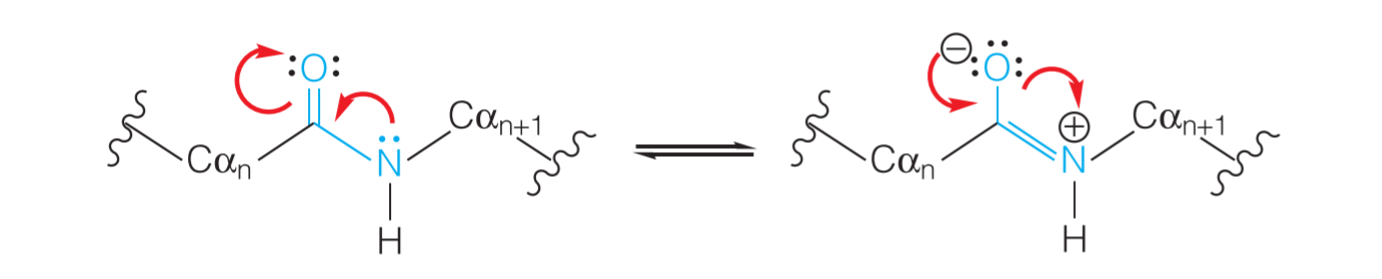
如上图所示,这六个原子常常是共平面的,并且可以形成两种形式的 resonance hybrid
2. Trans(反式) and Cis(顺式)
由于共平面的关系,肽键不能随意旋转,只能旋转180°,因此形成了trans和cis两种结构:
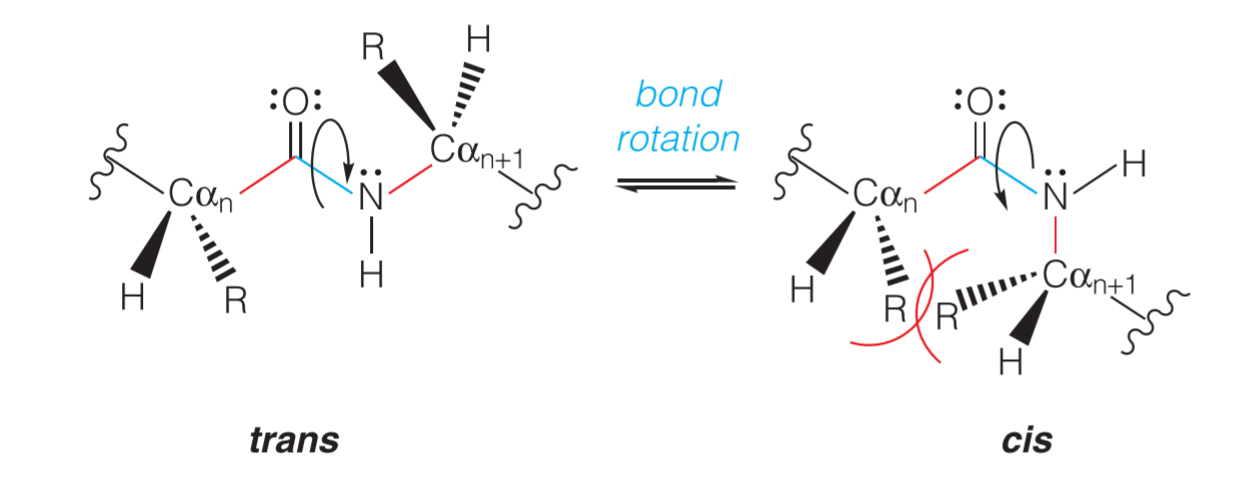
trans的结构更加常见(or say more favored),因为在cis结构里面,相邻氨基酸的R基团相邻更近,空间位阻(steric hindrance) 更大,因此更加不稳定
一个例外是X–Pro结构,因为脯氨酸的R基比较特殊,无论是trans还是cis,它们的空间位阻都比较大
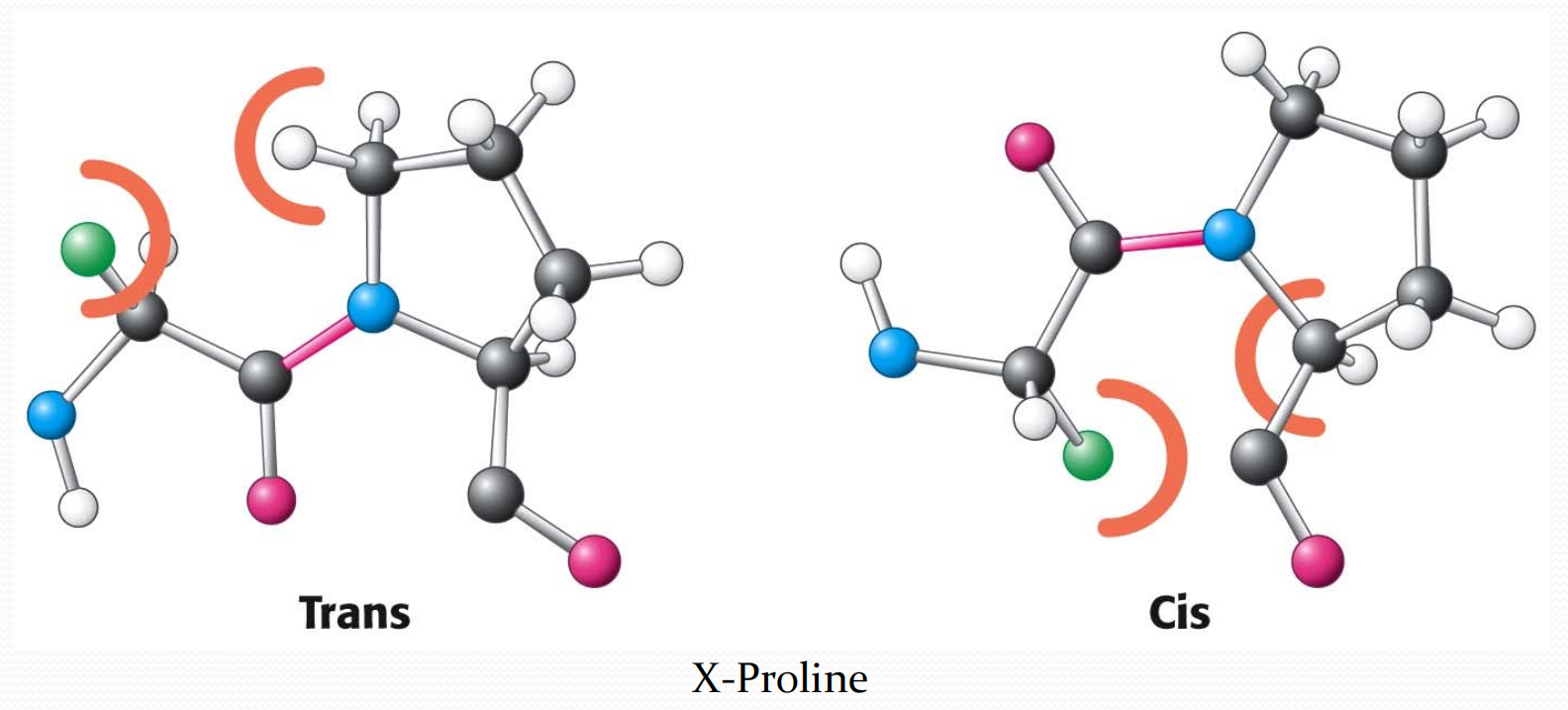
因此cis的比例相较于其他组合更高。但是trans与cis的比例仍然约为4:1.
3. Stability and Formation of the Peptide Bond
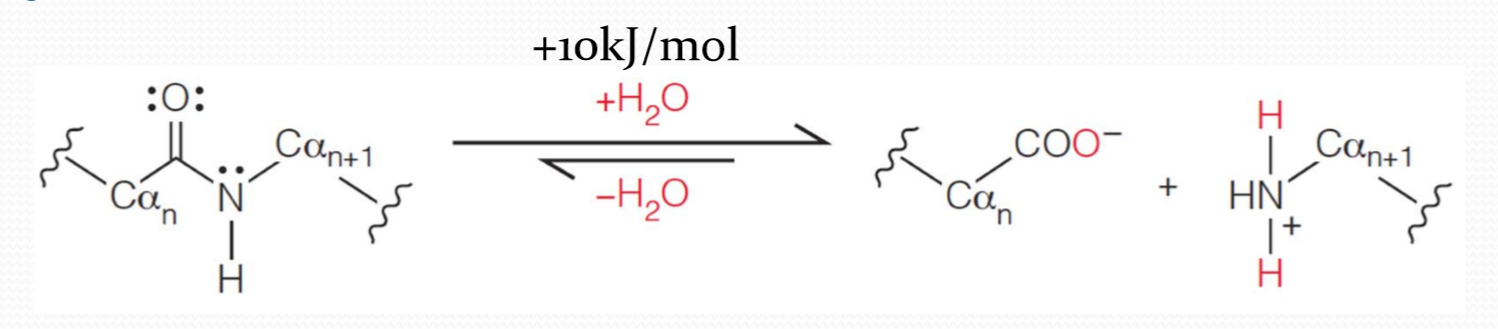
peptide bond could be formed by the elimination of a water molecule between two amino acids.
In fact, in an aqueous environment peptide bond formation is thermodynamically unfavored. The free energy change for peptide bond formation at room temperature in aqueous solution is about +10kJ/mol,therefore the favored reaction under these conditions is the hydrolysis(水解) of the peptide bond.
every amino acid must be activated by an ATP-driven reaction before it can be incorporated into proteins.
Peptide bond hydrolysis
- Strong mineral acid(无机酸) (e.g., 6 M HCl) cleaves all peptide bonds (including the Asn and Gln amide bonds)
- Chemicals that cleave at specific sites (e.g., CNBr(溴化氰, Cyanogen bromide) cleaves at Met)
- Proteolytic(蛋白水解的) enzymes (proteases,蛋白酶) that cleave at specific sites
4. dihedral angles 二面角
Conformation of a polypeptide chain can be solely described by $\phi$ and $\psi$ angles
Ramachandran plots of $\phi$ and $\psi$ show permissible angles for polypeptide chains
Some $\phi$ and $\psi$ angles are not allowed because of steric hindrance
Conformations of several types of secondary structures fall within permissible areas
The protein strand is -[-N–Cα–C]n-的不断重复,由于肽键所相关的六个原子共平面共享电子,因此Cα两旁存在两个平面,这两个平面可以以N–Cα或Cα–C为轴旋转,形成二面角
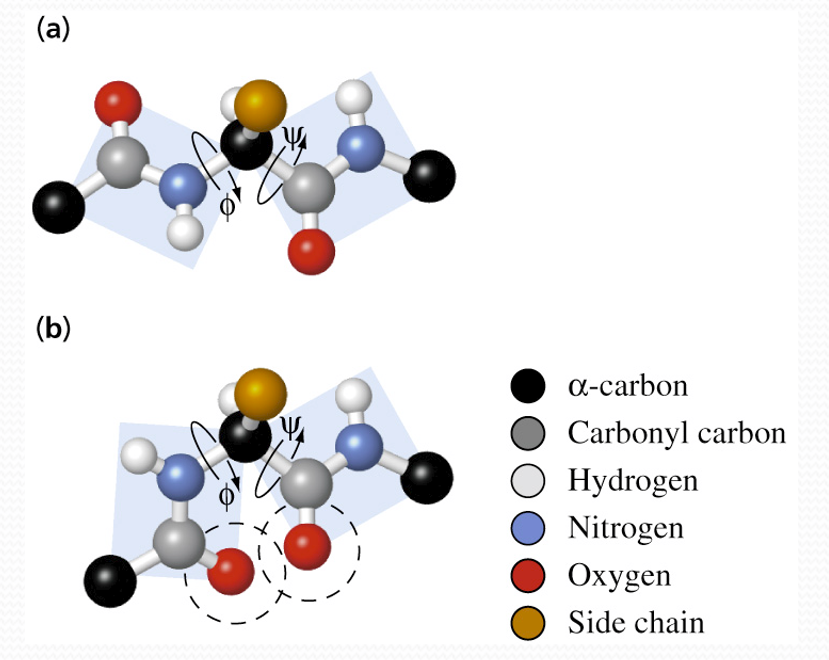
其中:
- $\phi$ (phi): Angle of rotation of N–C
αplane - **$\Psi$(psi): ** Angle of rotation of C
α–C plane
初始状态(phi与psi均为0°)
计算psi与phi的时候,需要定义它们何时为0°
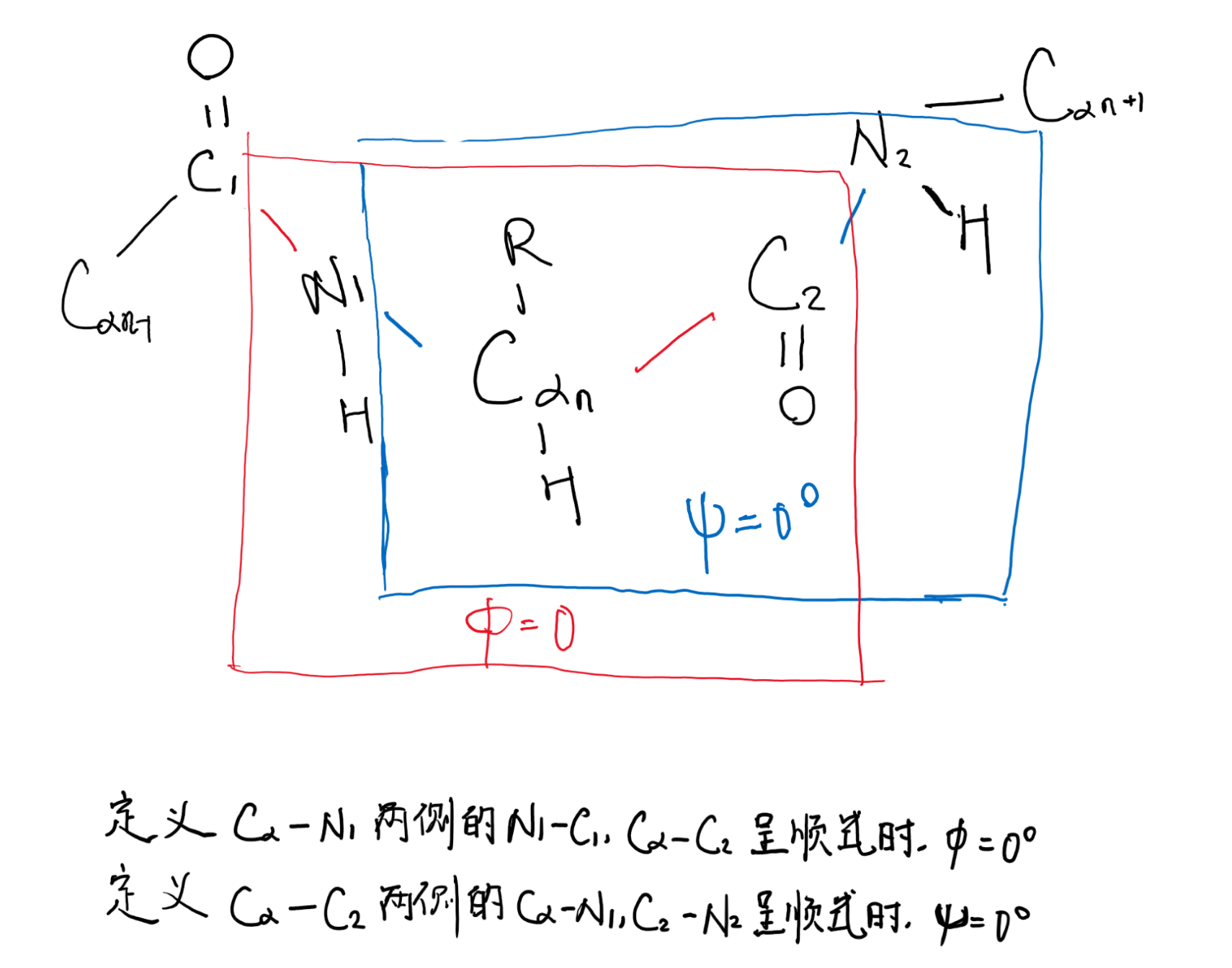
由Cαn向N1观察,顺时针旋转Cα—N1,得到的$\phi$角为正值,反之为负值
由Cαn向C2观察,顺时针旋转Cα—C2,得到的$\psi$角为正值,反之为负值
理论上psi和phi角可以是-180°~180°的任意值,但是由于主链上的原子和R group之间存在空间位阻(steric hindrance),因此有些角度是不允许的,例如phi和psi不为0°
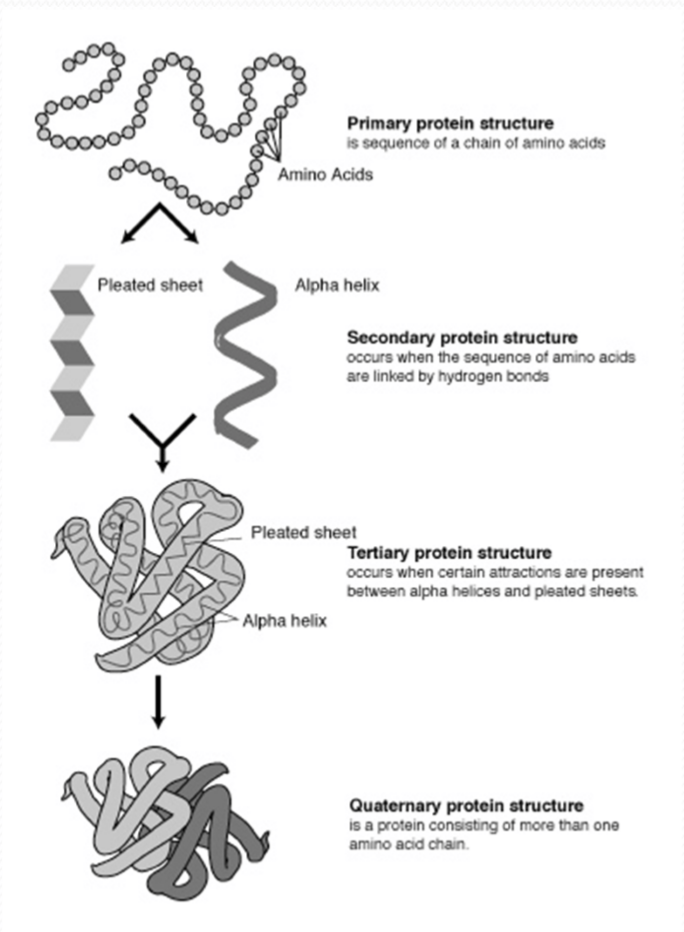
Four levels of protein structure
- Primary structure the amino acid sequence
- Secondary structure local folding into regularly repeating units
- Tertiary structure overall folding of a monomeric protein or subunit
- Quaternary structure subunit association
Primary Structure
1. The conformation of primary structure
Primary structure, the sequence of amino acids in a protein, is like the order of letters in a long word
Primary structure is determined by inherited genetic information
Each protein has its unique primary structure
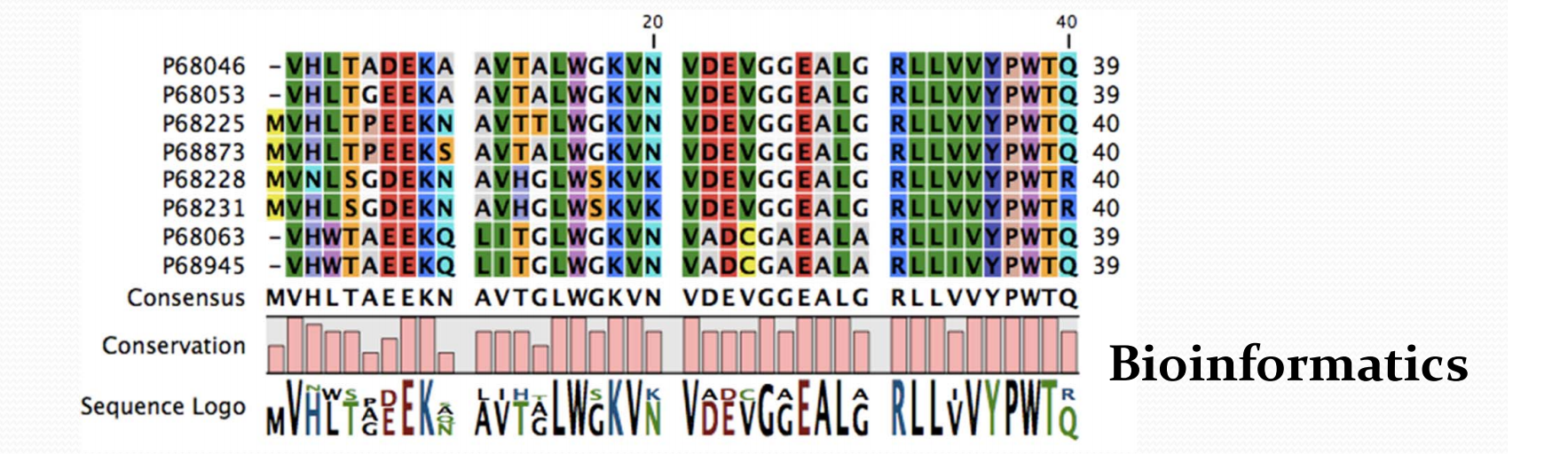
2. Nomenclature of protein sequence
- Residue number
- Sequence Homology
- Sequence identity (percentage of identical residues)
- Sequence similarity (percentage of similar residues)
- Amino acid conservation
Secondary structure
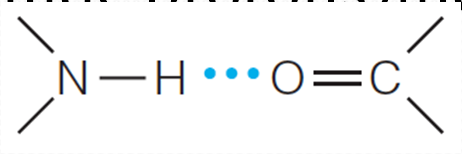
H-bond is the key factor of secondary structure.
Regular Ways to Fold the Polypeptide Chain
Regular Ways to Fold the Polypeptide Chain by Linus Pauling
The bond lengths and bond angles should be distorted as little as possible from those found through X-ray diffraction studies of amino acids and peptides.
No two atoms should approach one another more closely than is allowed by their van der Waals radii.
The amide group must remain planar and in the trans configuration. Consequently, rotation is possible only about the two bonds adjacent to the $\alpha$-carbon in each amino acid residue.
Some kind of noncovalent bonding is necessary to stabilize a regular folding. The most obvious possibility is hydrogen bonding between amide protons and carbonyl oxygens
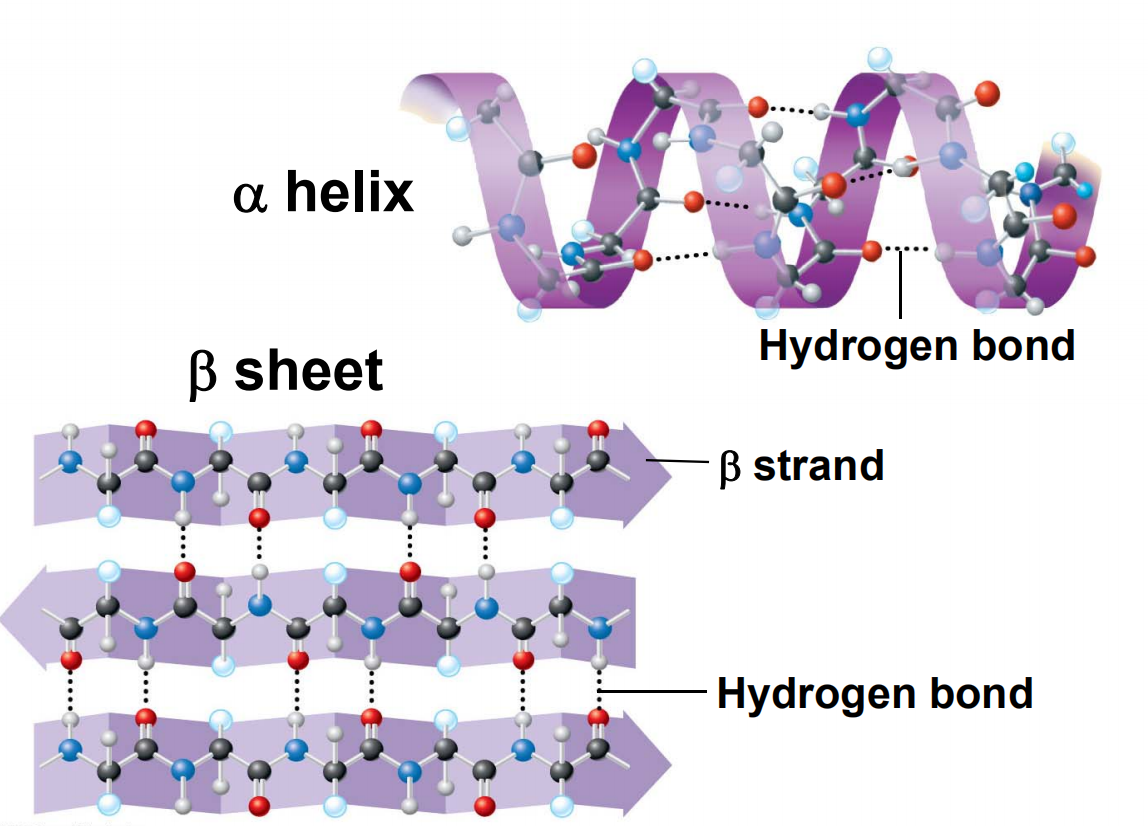

1. $\alpha$ Helix
Each C=O (residue n) forms a hydrogen bond with the amide hydrogen of residue n+4
3.6 amino acid residues per turn ($\frac{5.4}{1.5} = 3.6$)
Right‐handed
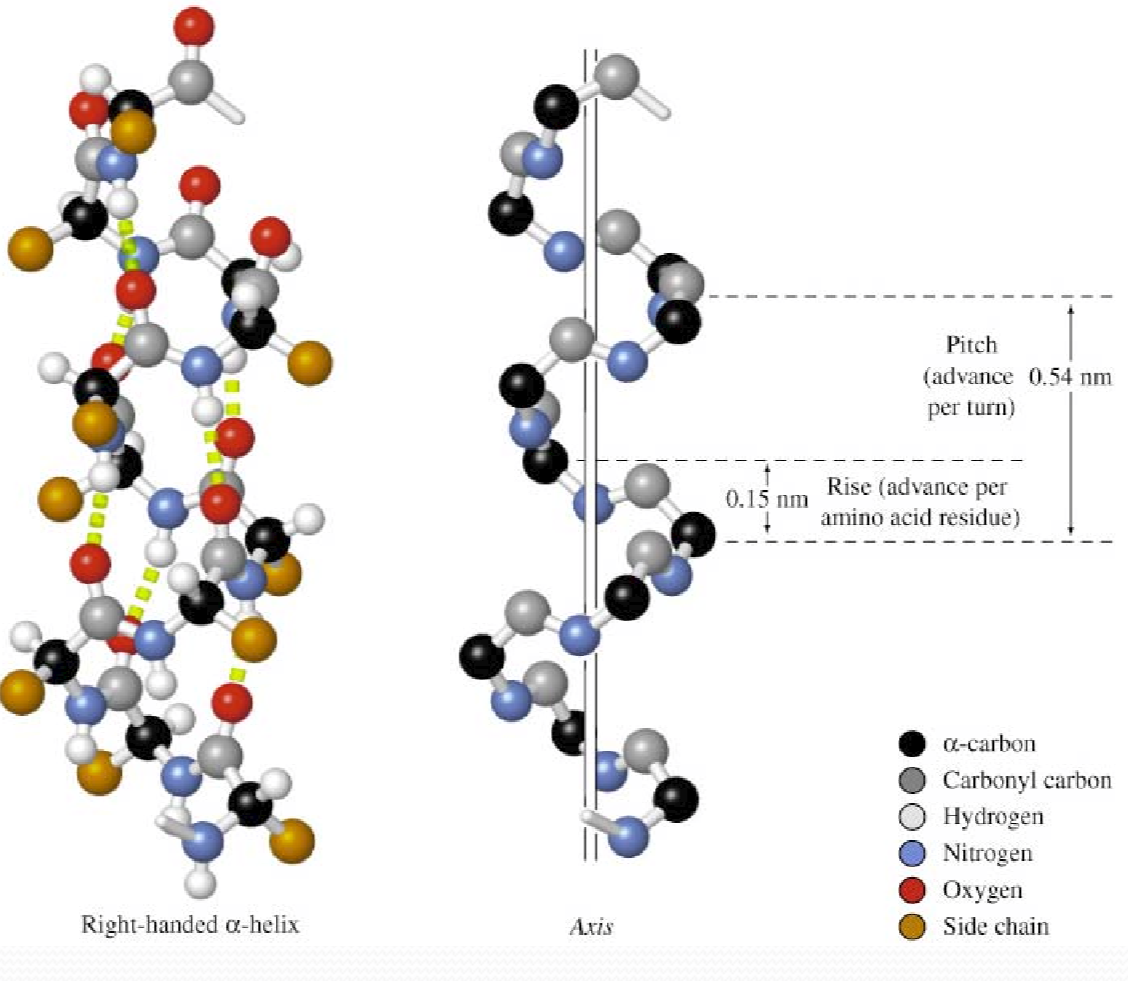
Axis
a helix has an axis that is a central line of symmetry running through it.
Pitch (p)
the spacing distance between individual adjacent coils of the helix.
For $\alpha$ helix, the p=5.4 Å/turn
Rise (h)
the distance the helix “rises” between adjacent polymer units.
<font color = “990000”>For $\alpha$ Helix, 1.5 Å/residue
Characteristics
- All side chains project outward from helix axis
- Side chains (C
α‐Cβ) point to the N‐terminus of helix - Carbonyl groups (C=O) point to the C‐terminus of helix
Other types of helices
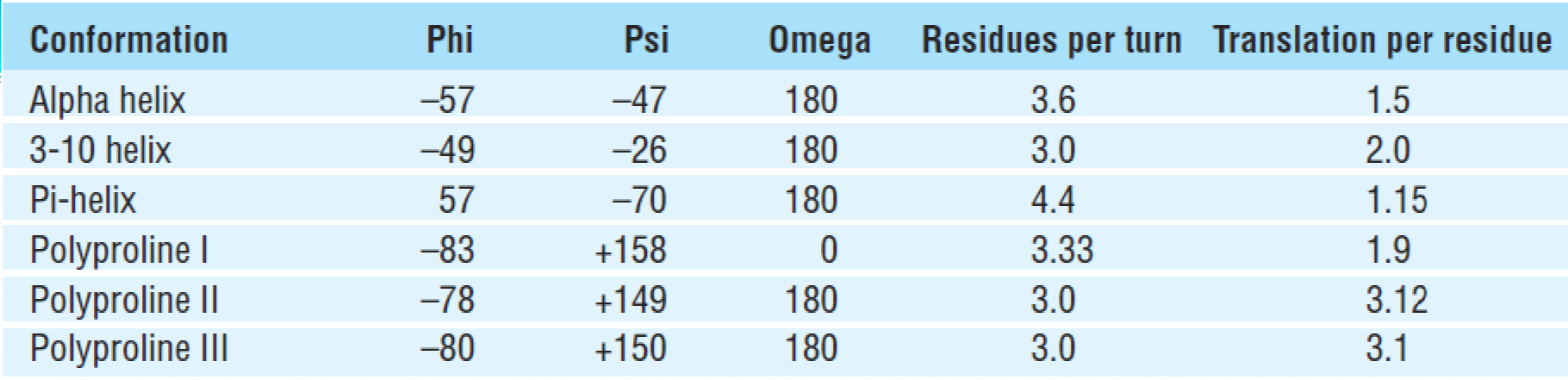
(1)3-10 helix
3-10 helix每圈的残基数 (n) 为3.0,每个肽键的C=O与前面的第2个肽基的N–H形成氢键,构成10圆环。
每个残基高度为0.2nm,螺距0.6nm,螺旋直径约为0.4nm。$\phi$ and $\psi$在-49°~26°左右
3-10 helix比α helix紧密,相邻螺圈中的C在一条直线上。但是由于3-10 helix中的氢键几何结构以及非键合相互作用并不处于最佳状态,所以在蛋白质结构中一般很短,通常作为α helix末端的最后一圈存在
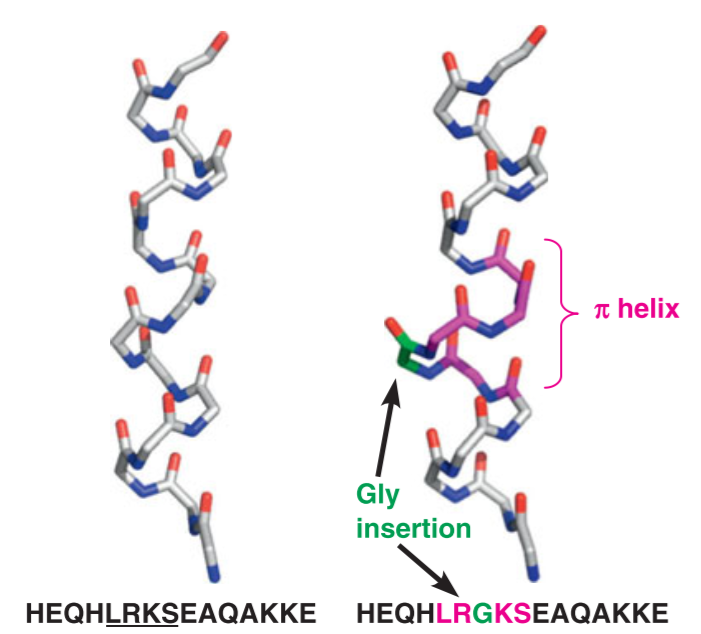
(2) $\pi$ helix
The $\pi$ helix is also called as an “$\alpha$ bulge” or “$\alpha$ aneurism” or “$\pi$ bulge”
pi-helix 的n = 4.4,h = 0.12nm,p = 0.52nm,螺圈直径约为0.6nm。
每个肽键的C=O与前面第四个肽基的N-H形成氢键,并构成16圆环。
由于$\pi$ helix 不稳定,因此在蛋白质结构中存在较少,但是这种结构的分布很广泛

$\pi$ helix的形成一般是一个aa的插入,使得$\alpha$ helix原有的H-bonding被打破,两个或多个残基与n+5的残基进行氢键链接
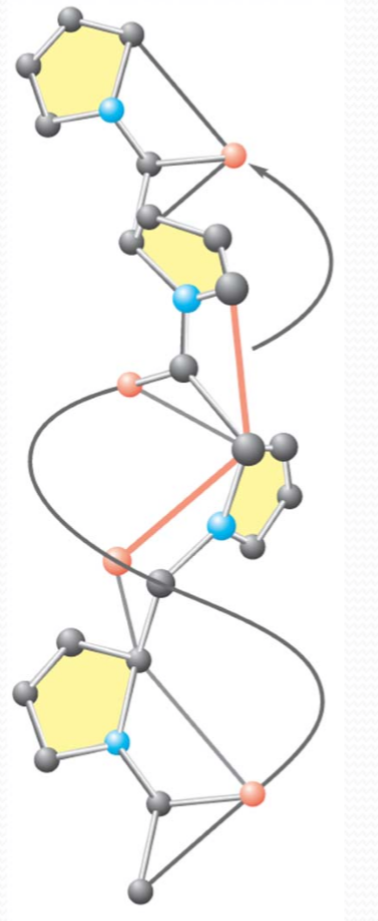
(3)Polyproline II helix
PolyprolineII helix
- Contains 1/3 proline residues
- No H‐bonds for main chain groups
- Left‐handed
- Polyglycinealso(聚甘氨酸) adopts this conformation
Helix dipole
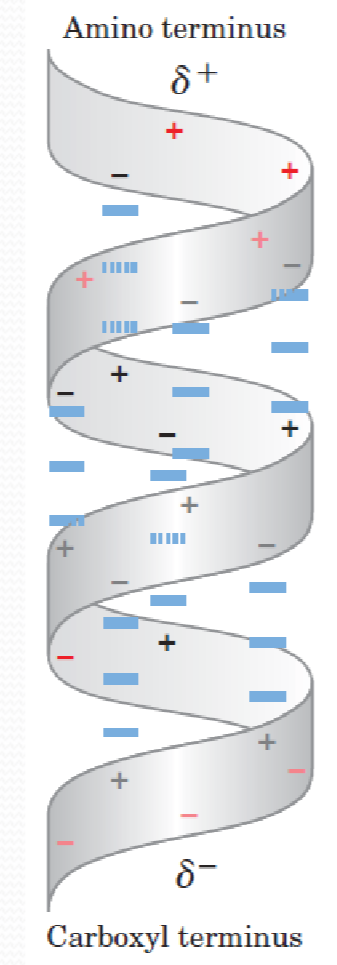
Aligned dipoles through hydrogen bonds (通过氢键排列的偶极子)
Increase with helix length
Negatively charged residues are located near the N‐terminus of the helix,N-terminus带正电
Positively charged residues are located near the C-terminus of the helix,C-terminus带负电
Constraints affect the helix formation
- Intrinsic(固有的) propensity of residues to form an helix
- The interactions between side chains
- Spaced 3‐4 residues apart
- The bulkiness(庞大) of adjacent side chains
- The interactions between residues at the ends of an helix
- The occurrence of Pro and Glyresidues:
- proline and gly rarely appears in helix:
- proline: no free -H to form H-bonds,proline一般出现在N-端,而不是middle或C-端
- glycine: no side chain,structure is flexible
2. β Strands and β Sheets
β Strands ‐polypeptide chains that are almost fully extended
β Sheets ‐multiple β strands arranged side‐by‐side
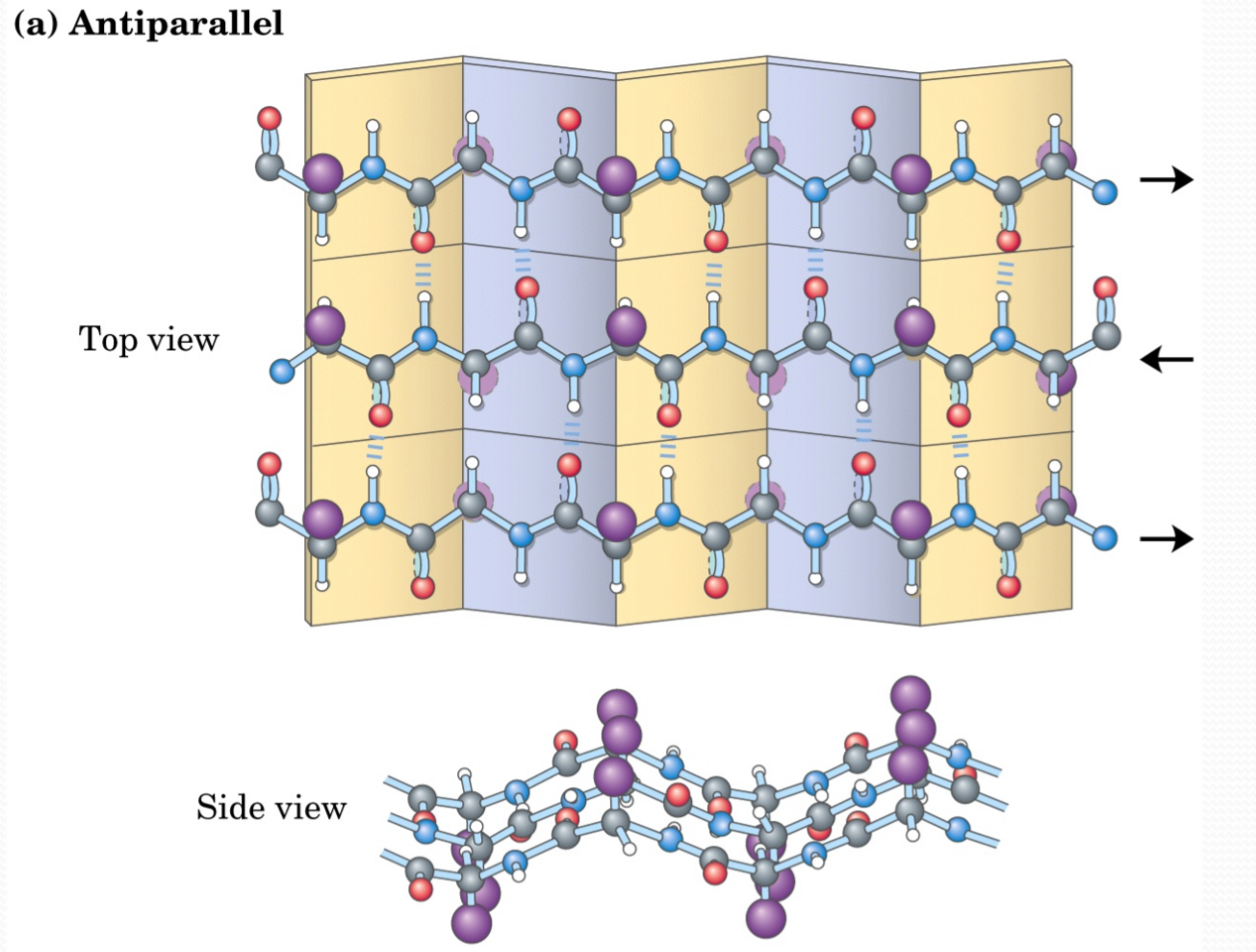
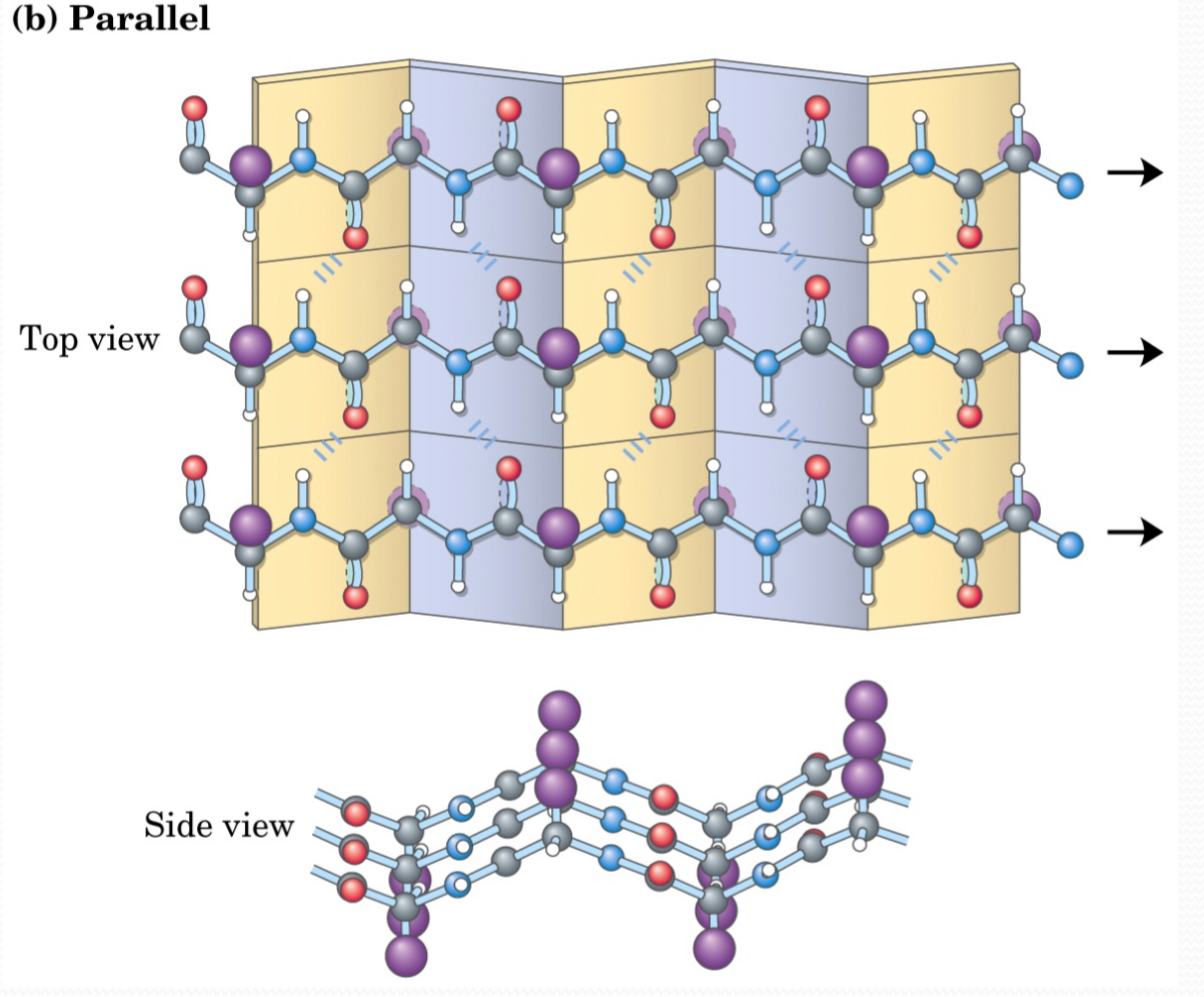
β Strands are stabilized by hydrogen bonds between C=O and ‐NH on adjacent strands
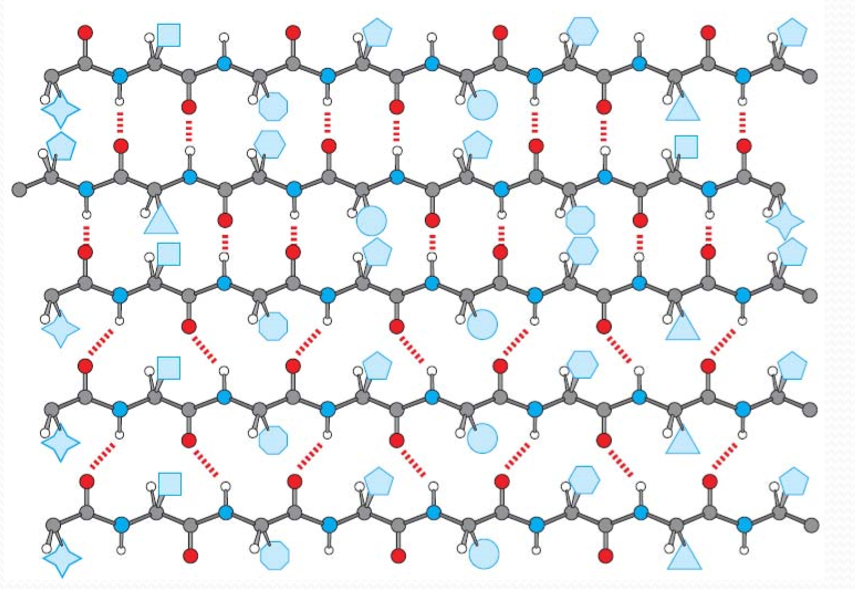
β Sheet side chains project alternately above and below the plane of the β strands
One surface of a β-sheet may consist of hydrophobic side chains that can interact with other hydrophobic residues in protein interior
Antiparallel β Sheets:
Parallel β Sheets:
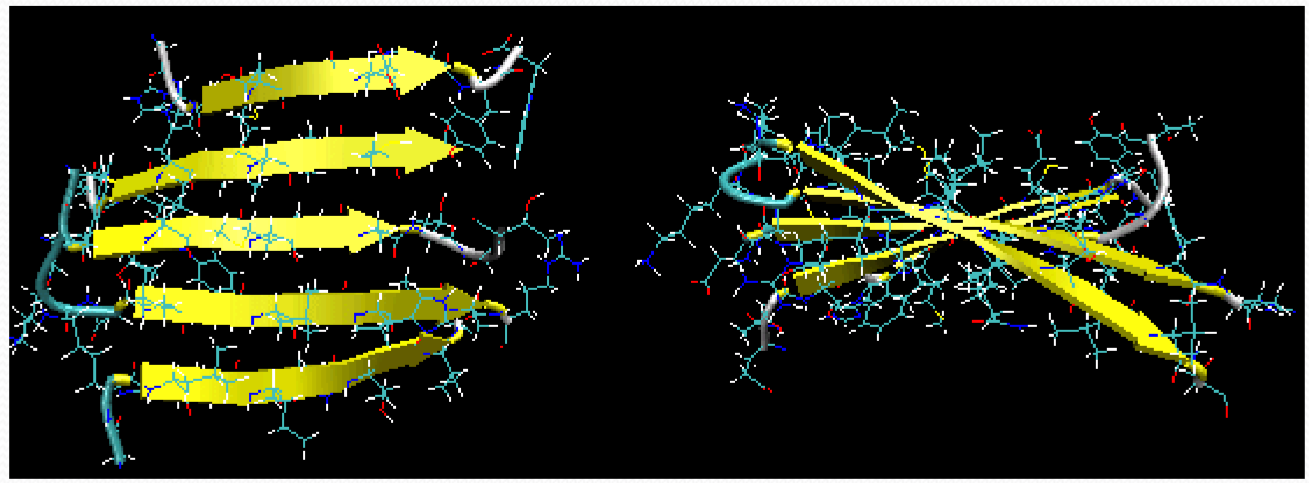
Twisted β Strand
The twist of the βsheet is due to the intrinsic twist of the polypeptide backbone.
3. Loops and Turns
Loops and turns connect helices and strands and allow a peptide chain to fold back on itself to make a compact structure
Loops ‐often contain hydrophilic residues and are found on protein surfaces
Turns ‐loops containing 5 residues or less
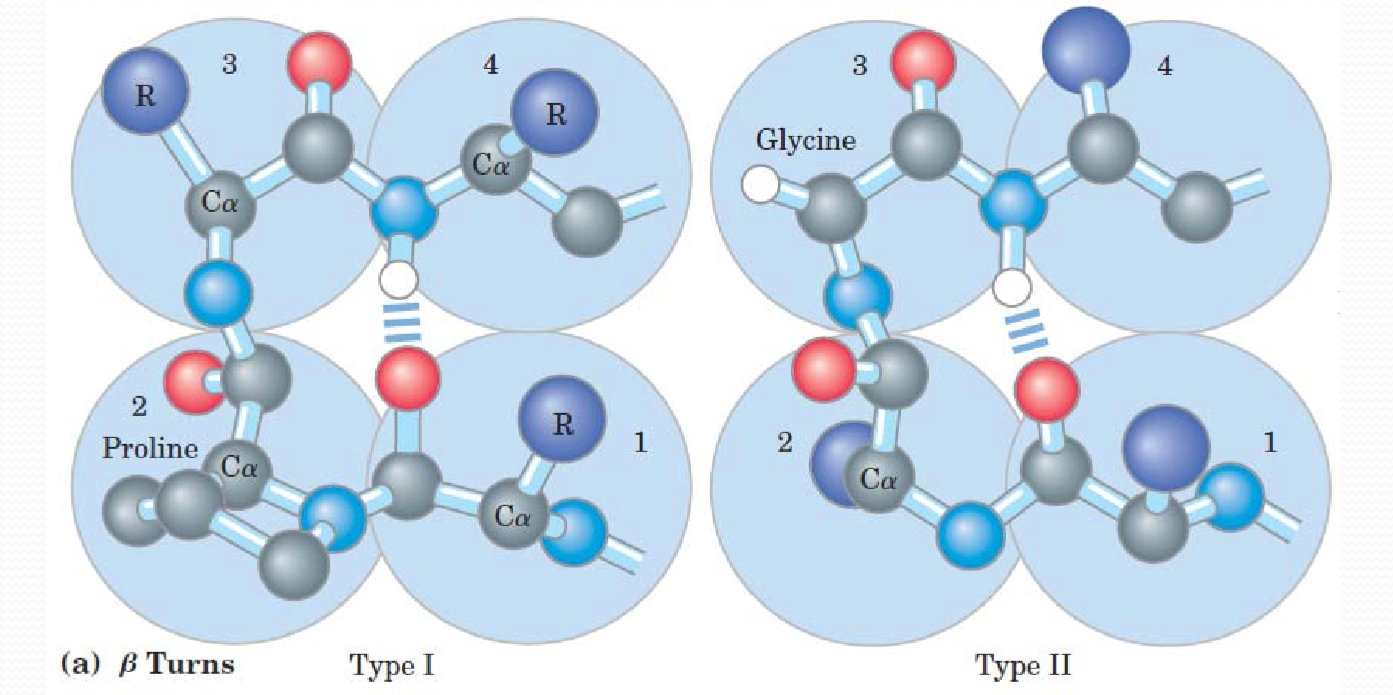
β turns
In beta turns, Type I involves proline, this type is less flexible. Type II involves glycine, more flexible.
4. Dihedral angles and Ramachandran plot
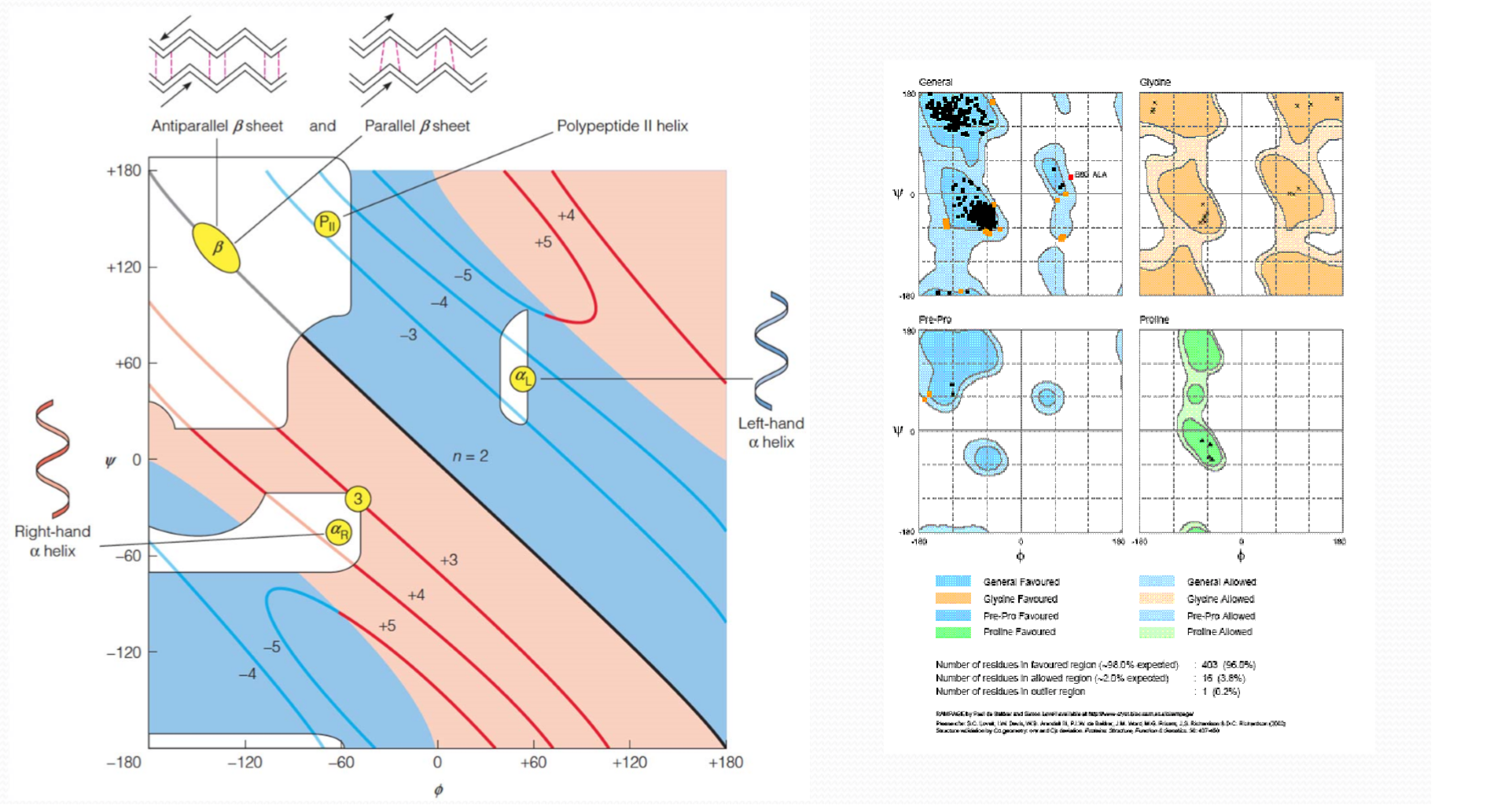 Ramachandran 图:表示了允许存在的(phi,psi)情况:
Ramachandran 图:表示了允许存在的(phi,psi)情况:
- 颜色深的more stable
- 颜色浅的allowed
- 无颜色的not allowed
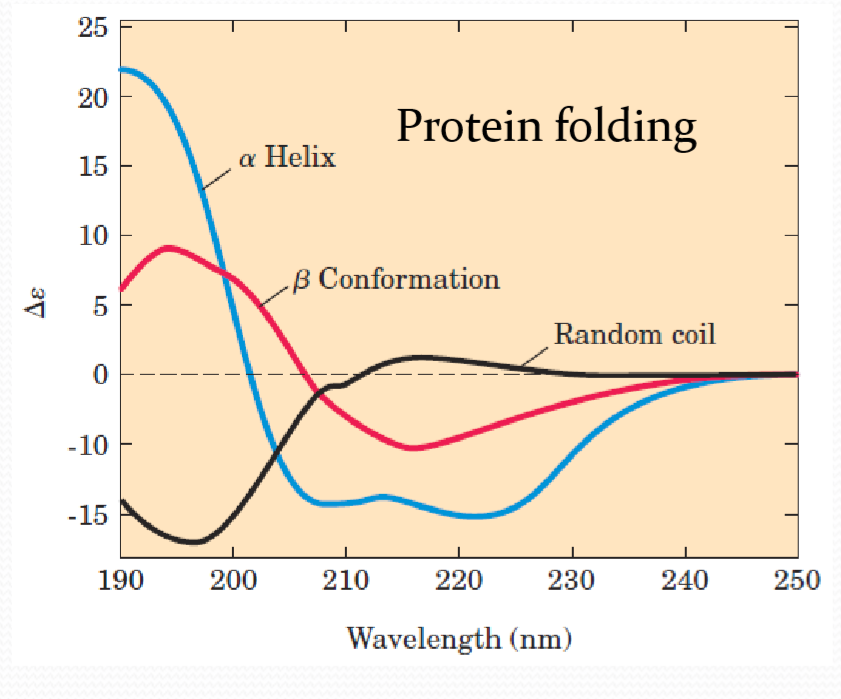
5. Circular Dichroism(CD) 圆二色性
一般而言,生物分子大多都具有手征性,也就是说它们在结构上有其他分子式相同但结构式不同的对映异构体(分为L型和D型),除此之外它们在光学特征上也有所差异。
通常光源在未经特殊处理前,其偏振方向通常是呈各种方向散射,经过起偏器偏振化后,光波会变为偏振方向单一的光波(称为线性偏振光)。当此平面偏振光通过手征性生物分子后,会分成左旋和右旋两道圆偏振光,最后再经过一道偏光镜使其重合为一线性偏光。
由于手征性生物分子结构上的影响,而使得左旋与右旋圆偏振光在折射率上有所差异,因此在重合后会产生附加的相位差,从而使得射出的合成线偏光在角度上产生偏转。
因为生物分子皆会在某一特殊光波长下有吸收光,它们除了对左旋与右旋的吸收度不同外,振幅也不同。因此随着时间的,左、右旋两道圆偏光重合后的行进方式将由原来的圆型变为椭圆型。由行进速度不同振幅也不同的左、右旋圆偏光叠加重合后所产生的不再是线性偏振光,而是椭圆偏振光,这种特性即称为圆二色性。
For proteins, spectra are obtained in the far UV region (190 to 250 nm) peptide bonds