L06 DNA Replication
一、DNA Replication
Requirement
DNA replication requires:
- Unraveling or relaxation of the supercoiled structure.
- Initiation of DNA replication at special sites. (Primer)
- Separation of the two strands from each other to create a single-stranded template.
- Replication of the two strands which run in an anti-parallel direction.
Segregation of Chromosome
1. Chromosome is a segregation device
A eukaryotic chromosome is held on the mitotic spindle by the attachment of microtubules to the kinetochore that forms in its centromeric region.
microtubule organizing center (MTOC) – A region from which microtubules emanate(发散).
- In animal cells the centrosome is the major microtubule organizing center, but plants use the nuclear envelope itself as the MTOCs.
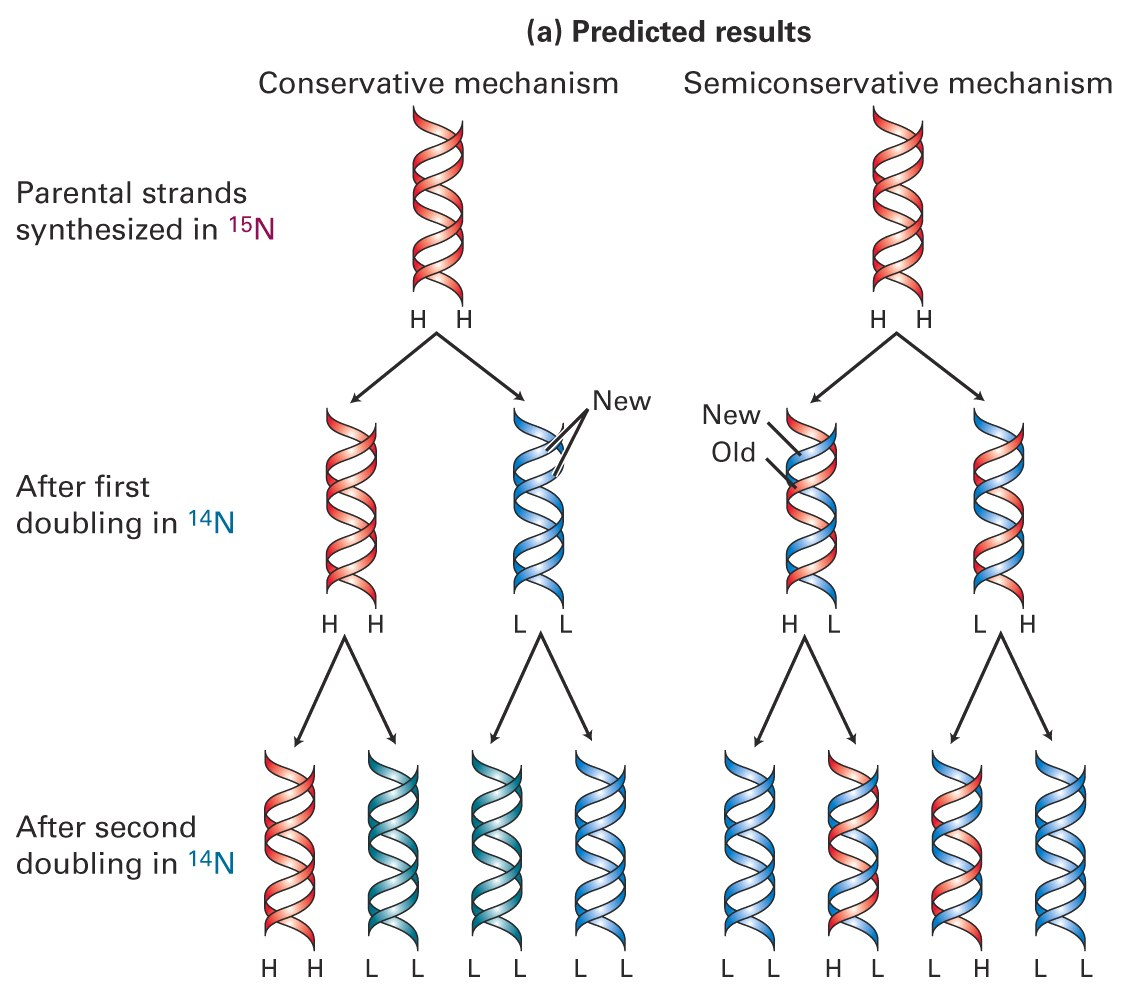
2. Semiconservative Nature (Meselson-Stahl Experiment)
The Meselson-Stahl Experiment proves the semi-conservative replication of DNA
Results matched prediction for semi-conservative replication;
- e.g. mixed strands were created (50% heavy, 50% light).
The Experiment Procedure
E. coli cells grown in ,heavy” nitrogen to label cellular DNA. (‘H’ in
figure)
Transferred to normal “light” nitrogen, grown, and then DNA separated by density-gradient centrifugation.
If replication were ‘conservative (at left), then after replication, only old and new types are present
If semi-conservative, after replication, mixed strands would be created (50% heavy, 50% light).
3. Theta Structures introduced in the circular DNA Replication
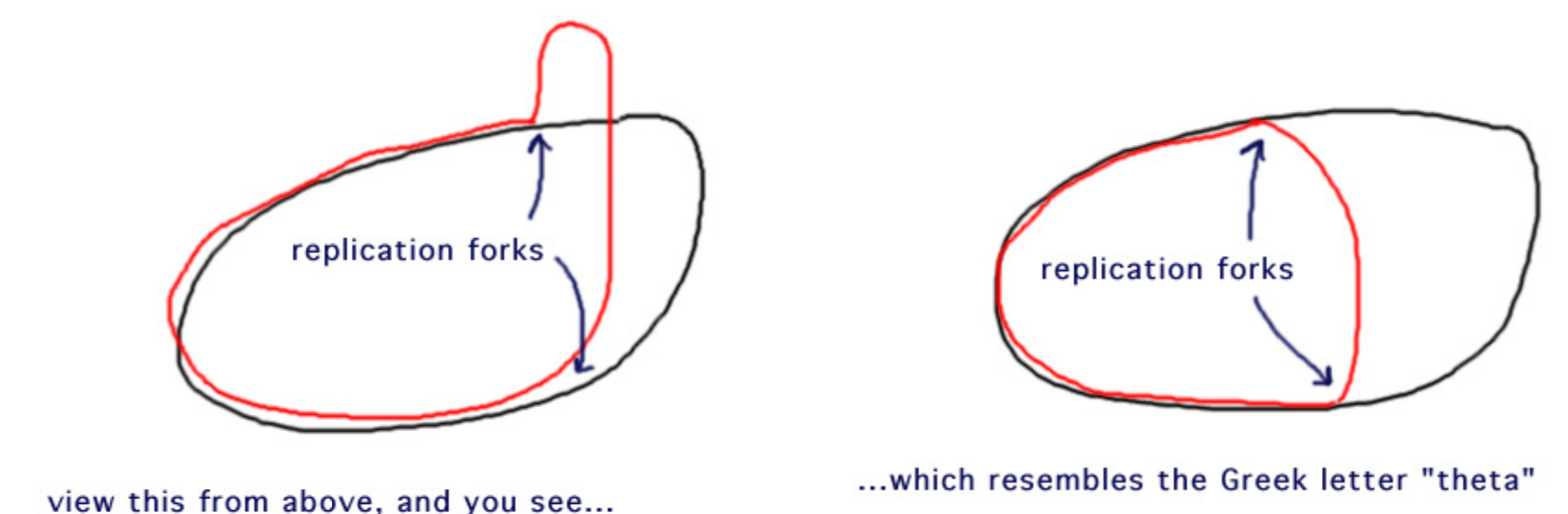
In the absence of swiveling(转动的), the unwinding(解退) DNA in one region would cause tightening or over-winding of the other strand.
Relaxation of this stress is performed by topoisomerase enzymes.
By cutting one strand of DNA, these enzymes allow one DNA to swivel around the other.
Direction and Progress
1. Direction
Direction for one strand
The DNA replication has a direction from 5’ to 3’
Direction for the whole double-strand DNA
所以复制从一点开始,对于一条double-strand DNA来说同时向两边扩展
The replicating strands form a “bubble”.
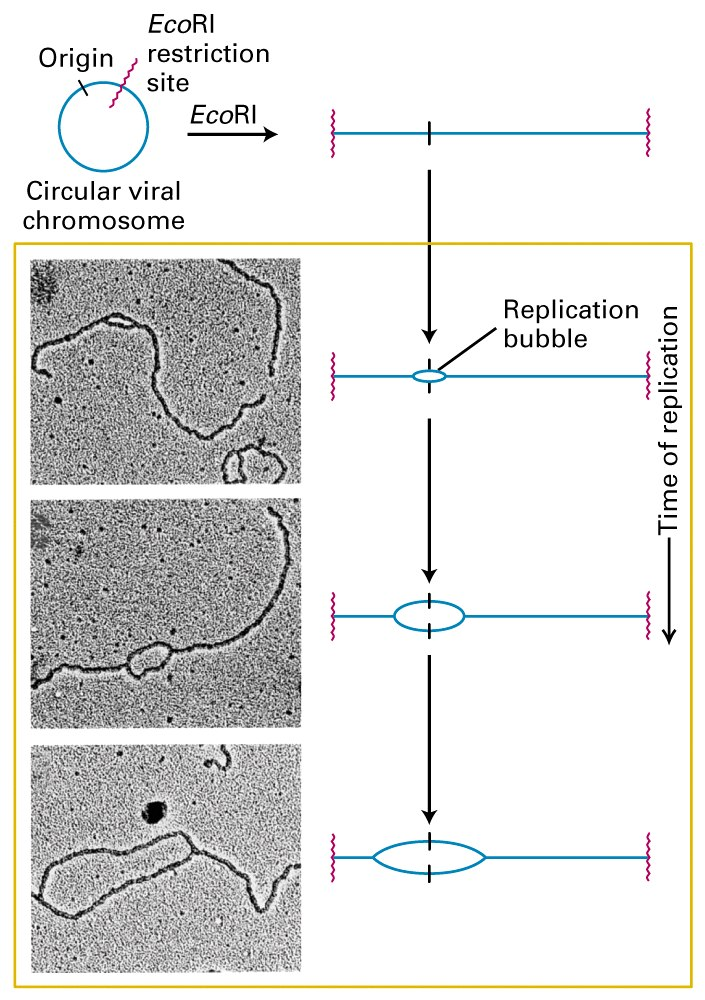
The two sites of replication are proceeding identically, with a “fork” formed from the parent strand to the new parent/daughter hybrid strands.
Bidirectional(双向的) strand growth is universal to prokaryotes and eukaryotes.
2. dNTPs
The template stand directs which of the four dNTPs are added in the extension of the 3’ end.
3. Primes
DNA is formed from dNTP monomers; synthesis proceeds in the 5’ to 3’ direction. DNA polymerases require a primers such as a short piece of RNA.

4. Semidiscontinuous DNA Replication
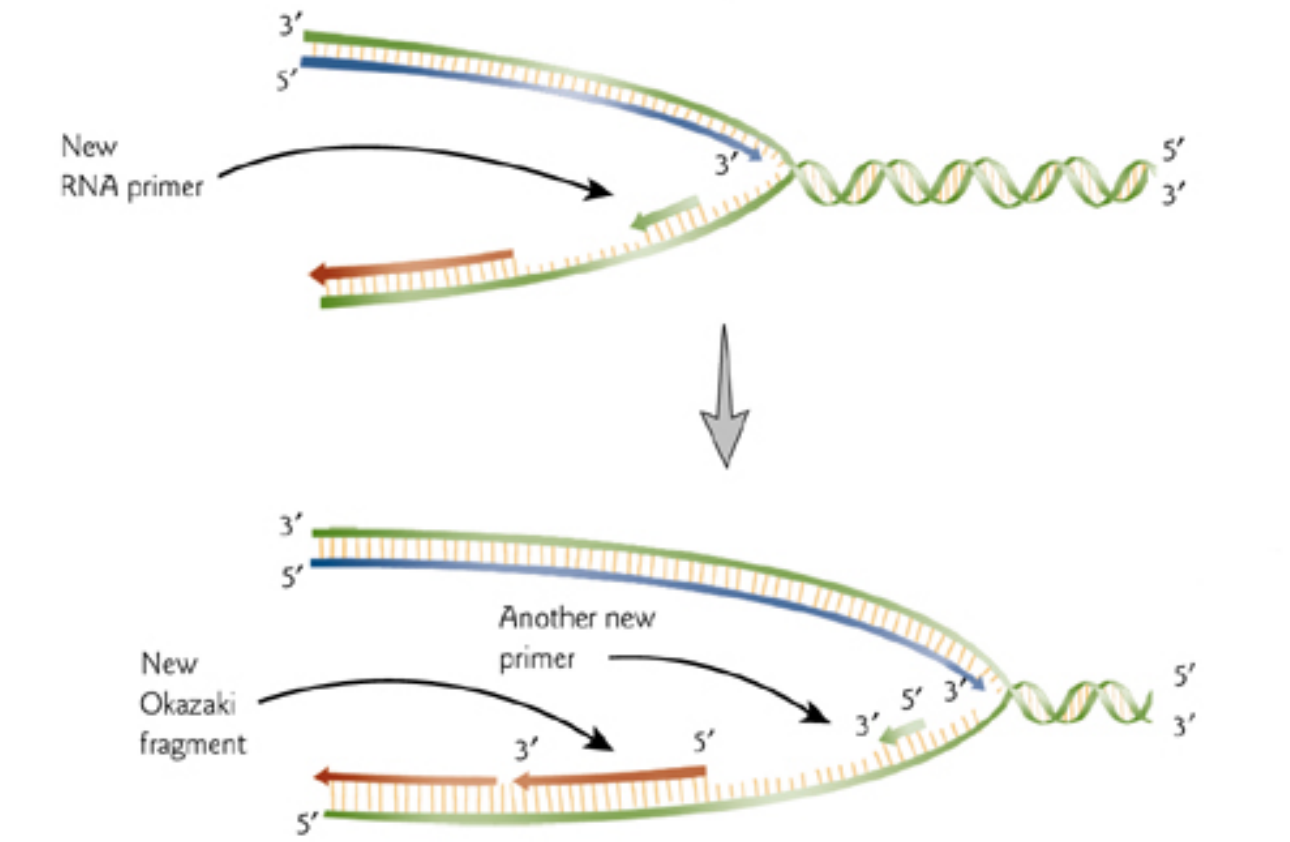
DNA is formed from dNTP monomers; synthesis proceeds in the 5’ to 3’ direction.
DNA polymerases require a primers such as a short piece of RNA.
5. DNA Replication Forks
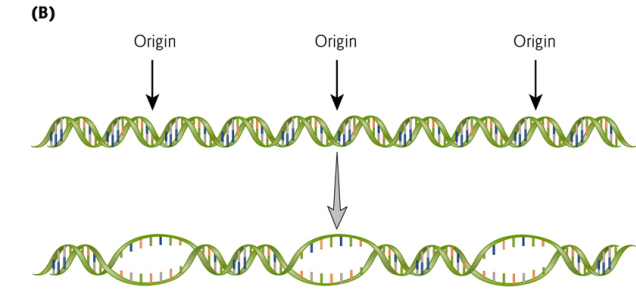
DNA replication forks occur at many locations, these finally merge to create the completely replicated molecules
6. Bidirectional(双向) Replication
The replicating strands form a “bubble”.
The two sites of replication are proceeding identically, with a “fork” formed from the parent strand to the new parent/daughter hybrid strands.
Bidirectional strand growth is universal to prokaryotes and eukaryotes.
7. Replication Initation Point Mapping
Newly synthesized DNA is used as template.
Primer extension stops at DNA-RNA junctions on the nascent strand because the DNA polymerase included in the reaction cannot use RNA as a template.
TP = transition point, location of shift from discontinuous to continuous synthesis.
DNA polymerase
DNA polymerases often have a 3′–5′ exonuclease activity that is used to excise(切除) incorrectly paired bases.
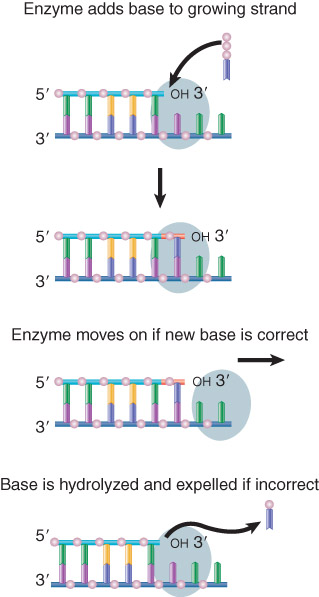
Bacterial DNA polymerases scrutinize the base pair at the end of the growing chain and excise the nucleotide added in the case of a misfit.
1. holoenzyme 全酶
The DNA polymerase holoenzyme consists of subcomplexes
A clamp loader places the processivity subunits on DNA, where they form a circular clamp around the nucleic acid.
One catalytic core is associated with each template strand.
Untwisting of Supercoiled DNA
The untwisting of DNA is needed for DNA replication.
This localized unwinding creates tension further along the molecule, particularly in a circular DNA molecule in which the tension cannot be released at the ends.
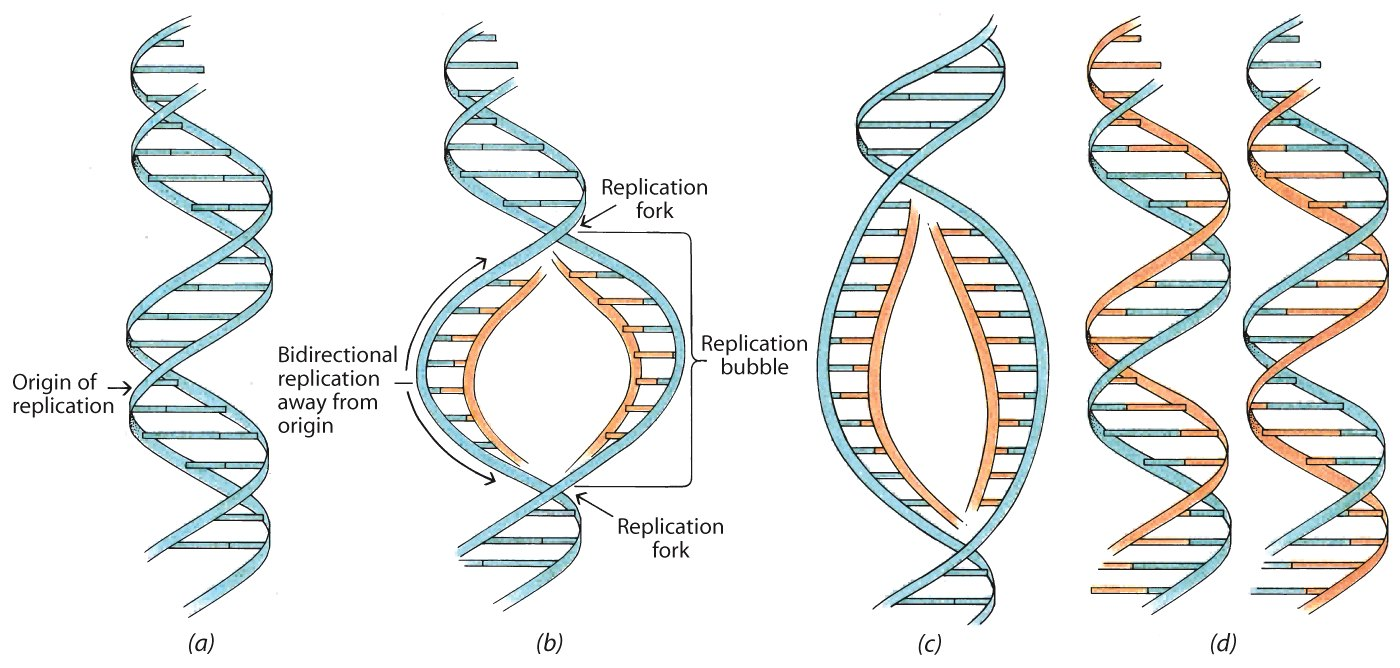
Characteristic Structures or Molecules
1. Origins of Replication
Origins of Replication differs in Prokaryotic and Eukaryotic cells
Single origin for bacterial chromosomes
Many origins for eukaryotic chromosomes, multiple replication forks.
2. Origin Reading Complex, ORC
Histone removal at the site of replication to allow access to DNA.
Histone deposition, with nucleosomes reassembled on nascent(初期的,开始存在的) DNA via interaction with chromatin assembly factor.
Prereplication complex formation at the origin by the association of the origin recognition complex(ORC).
Replication licensing
Once the ORC binds, additional proteins are recruited(ORC结合后,会吸引结合更多的蛋白质)
The Assembly of Prereplication Complex
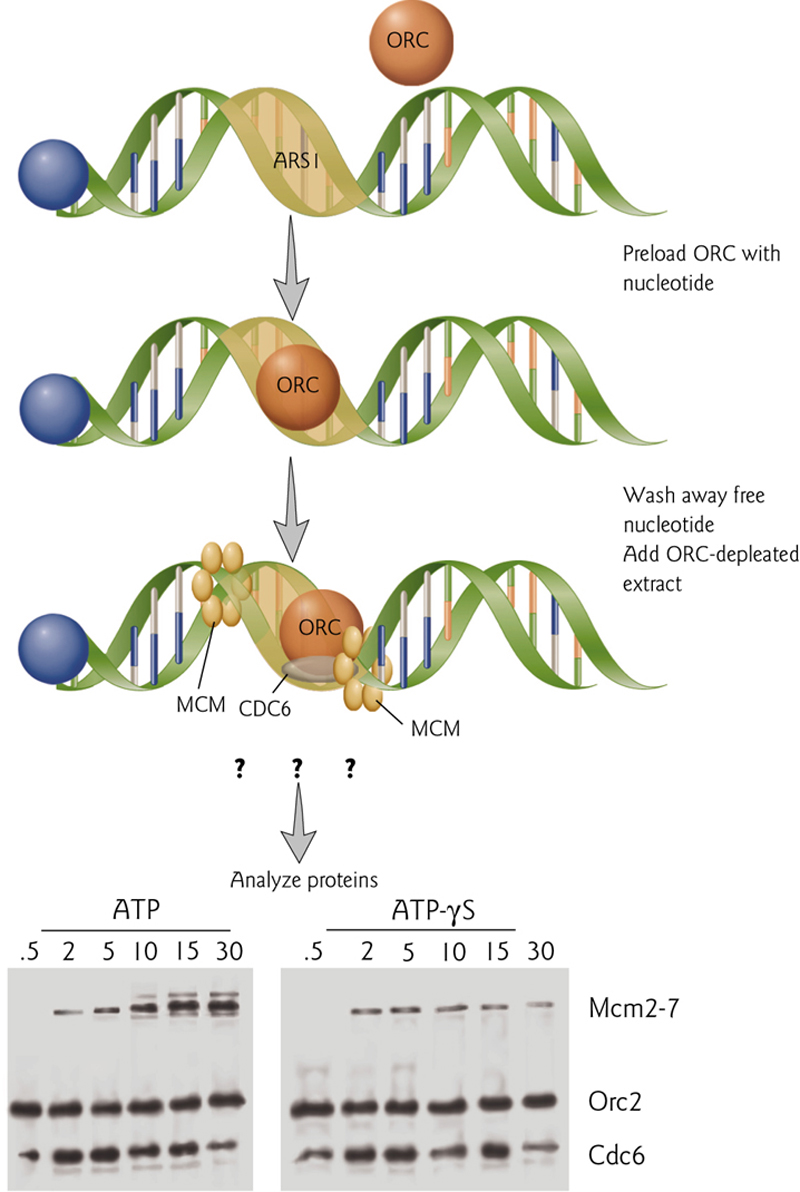
ATP hydrolysis by the origin recognition complex (ORC) stimulates prereplication complex assembly, which is inhibited by ATP-γS-bound ORC (a non-hydrolyzable form of ATP).
3. Helicase(解旋酶) and Topoisomerase
- Helicase initiates local melting/unwinding of DNA at “origin of replication” (ori).
- Single-strand binding proteins stabilize strands
- Primase (RNA polymerase) forms short RNA primer.
- DNA polymerase uses RNA primer to continue synthesis (switch of polymerases).
- Topoisomerase relieves supercoils.
4. Primase(RNA polymerase) and RNA primer
Single-strand binding proteins stabilize strands
- Primase (RNA polymerase) forms short RNA primer.
- DNA polymerase uses RNA primer to continue synthesis (switch of polymerases).
5. Lagging Strand – Leading Strand and Okazaki Fragment
On lagging strand, ‘Okazaki fragments’ are started every few hundred bases.
When one Okazaki fragment meets RNA primer of next strand, RNA is removed and DNA strands are ligated together by ‘ligase’.
Filing in of gaps left by removal of RNA primers
- Joining of Okazaki fragments by DNA ligase I (DNA 连接酶 I)
- Human DNA Ligase I encircles(环绕)DNA during ligation
6. Proliferating cell nuclear antigen 增殖细胞核抗原(PCNA)
“Proliferating cell nuclear antigen” (PCNA) originally identified in dividing cells. Interacts with many other proteins.
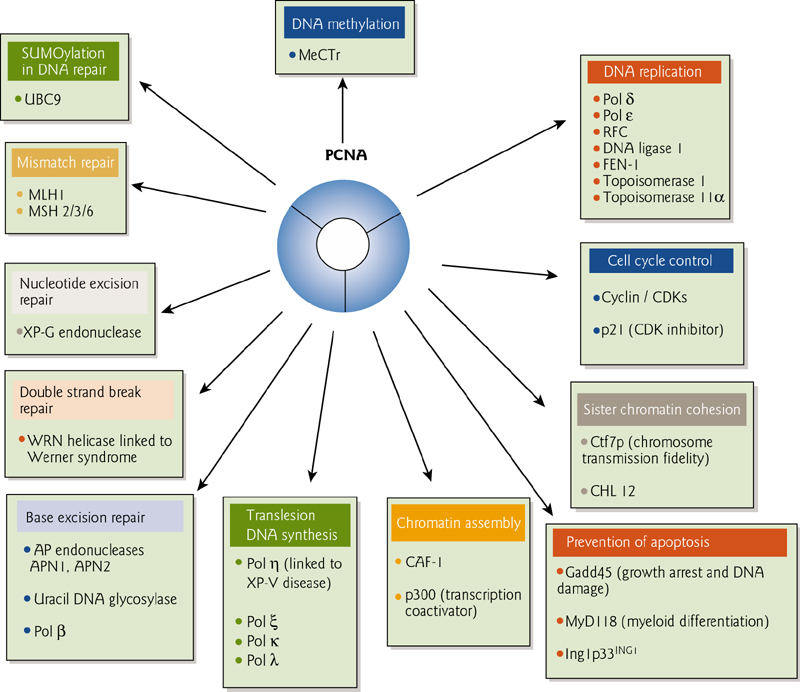
PCNA is a “sliding clamp” that encircles DNA and allows proteins to stay adhered to DNA during processing.
7. DNA Ligase
- Joining of Okazaki fragments by DNA ligase I (DNA 连接酶 I)
- Human DNA Ligase I encircles(环绕)DNA during ligation
The ends of linear DNA are a problem for replication
Special arrangements must be made to replicate the DNA strand with a 5’ end
DNA End Replication Problem
Because of the requirement for a primer on the lagging strand, when the final primer is removed, this leaves an 8-12 nt region uncopied at the 5’ end.
Terminal proteins enable initiation at the ends of viral DNAs
Activation of DNA replication
1. CDK1
Origin of replication ativated once during the cell cycle
DNA synthesis is restricted to only the S (‘synthesis’) phase of the cell cycle.
Replication is therefore carefully regulated.
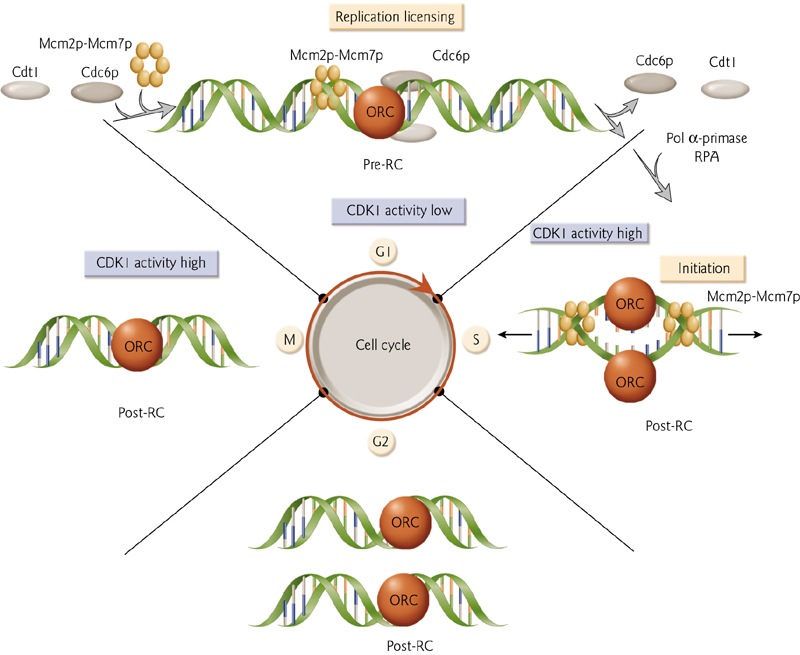
2. Assembly of The Prereplication Complex
The data suggest that ATP hydrolysis by the origin recognition complex (ORC) stimulates prereplication complex assembly, which is inhibited by AFR-vS-bound ORC (a non-hydrolyzable form of AFP).
Assembly of Nucleosomes
Nucleosome assembly after DNA replication
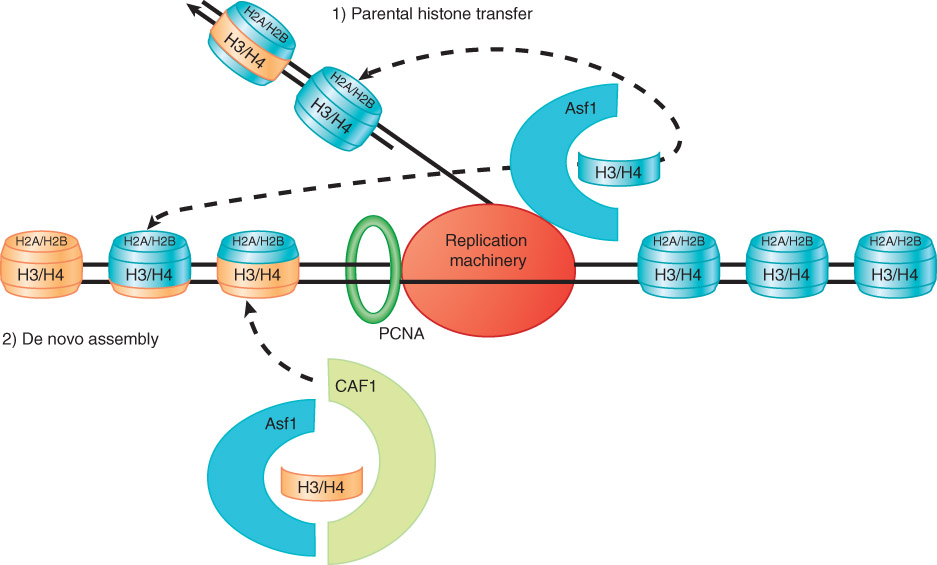
Replication fork passage displaces histone octamers from DNA.
Histones may be added back to the DNA strands as either entire new tetramers, or mixed pairs of old and new dimers.
Simple Biological System – Rolling Cycle
Simple Biological system use “Rolling Circle” or “Covalent extension” Replication
This produces large amounts of DNA, which is a “concatamer(串联体)” of many molecules of the same template.
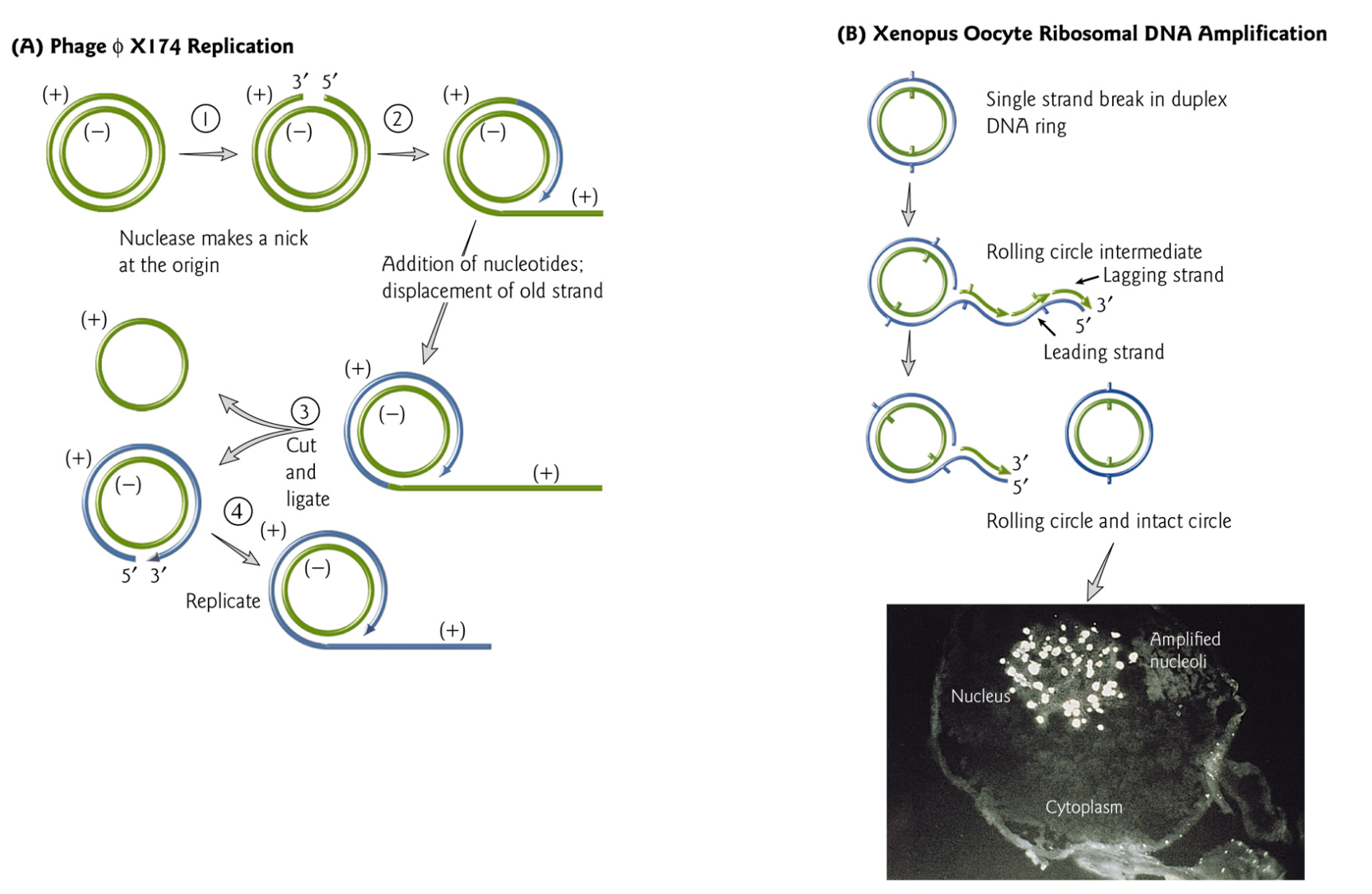
Telomere
A telomere consists of a simple repeat where a C+A-rich strand has the sequence $C_{>1}(A/T)_{1-4}$
A typical telomere has a simple repeating structure with a G-T-rich strand that extends beyond the C-A-rich strand.
Telomere have simple repeat sequences which seal the chromosome ends
Mutation in telomerase causes telomeres to shorten in each cell division. Eventual loss of the telomere causes chromosome breaks and rearrangements.
The telomere is required for the stability of the chromosome end.
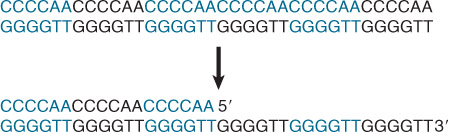
1. Telomerase
Telomerase puts the ends of eukaryotic chromosomes
Telomerase uses the 3′-OH of the G+T telomeric strand to prime synthesis of tandem TTGGGG repeats.
The RNA component of telomerase has a sequence that pairs with the C+A-rich repeats.
One of the protein subunits is a reverse transcriptase that uses the RNA as template to synthesis the G+T-rich sequence.
RNA in telomerase pairs with DNA and polymerase extends DNA at 3’ end, which can then be filled in either by lagging strand synthesis or T-loop formation.
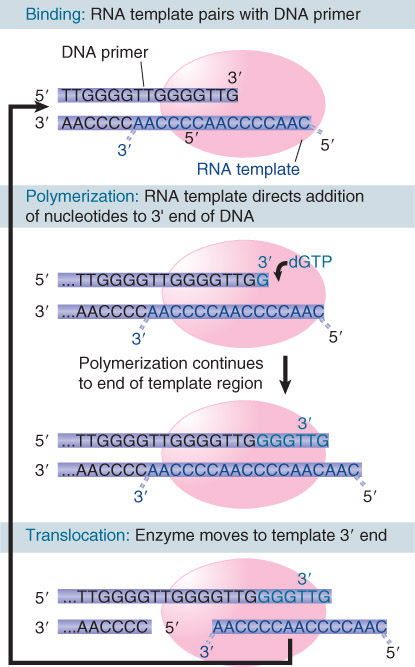
2. TRF2
The protein TRF2 catalyzes a reaction in which the 3′ repeating unit of the G+T-rich strand forms a loop by displacing its homolog in an upstream region of the telomere.
The 3’ single-stranded end of the telomere (TTAGGG)n displaces the homologous repeats from duplex DNA to form a t-loop. The reaction is catalyzed byTRF2.
3. The length of telomeres
The length of the telomere is determined by concentration of the POT1 (Protection of Telomeres1) protein bound to the DNA.
This is sensed by the telomerase and the level either represses or activates telomerase activity.
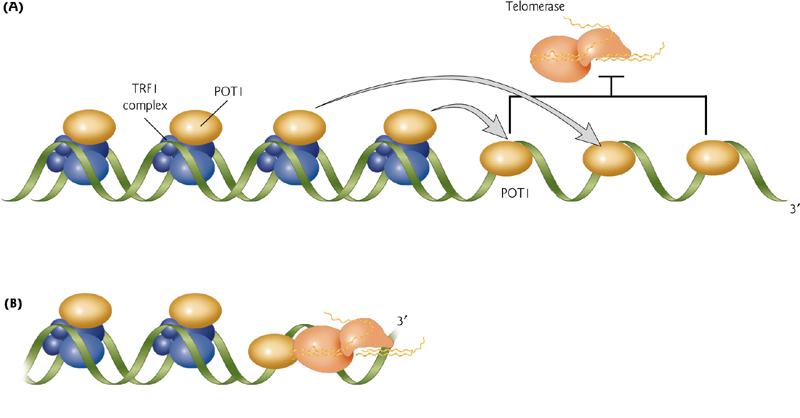
4. The existence of telomerases
Telomerase is expressed in actively dividing cells and is not expressed in differentiated cells.
Loss of telomeres results in senescence(衰老).
Escape from senescence can occur if telomerase is reactivated, or via unequal homologous recombination to restore telomeres.
二、DNA Proofreading
DNA polymerase I
DNA polymerase I has a 3’ to 5’ exonuclease activity to remove erroneous bases. The normal replication direction is 5’ to 3’.
The DNA polymerase I functions usually from 3’ to 5’
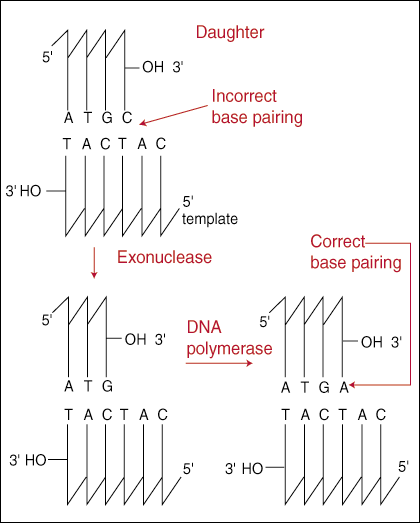
DNA polymerase I has a 3’ to 5’ exonuclease activity to remove erroneous bases. The normal replication direction is 5’ to 3’.