L10 Forward And Reverse Genetics
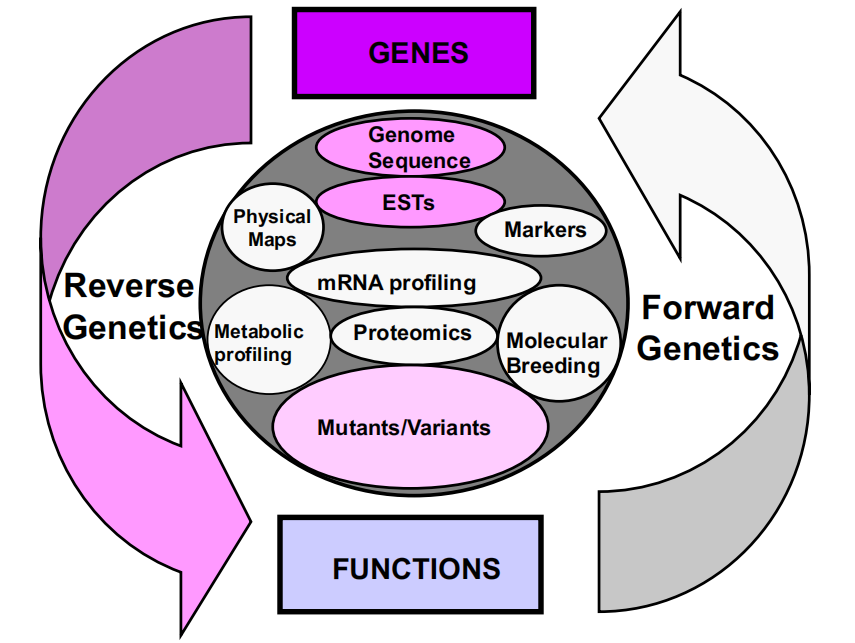
Two ways to study genes and their functions
一、The Forward Genetics Approach
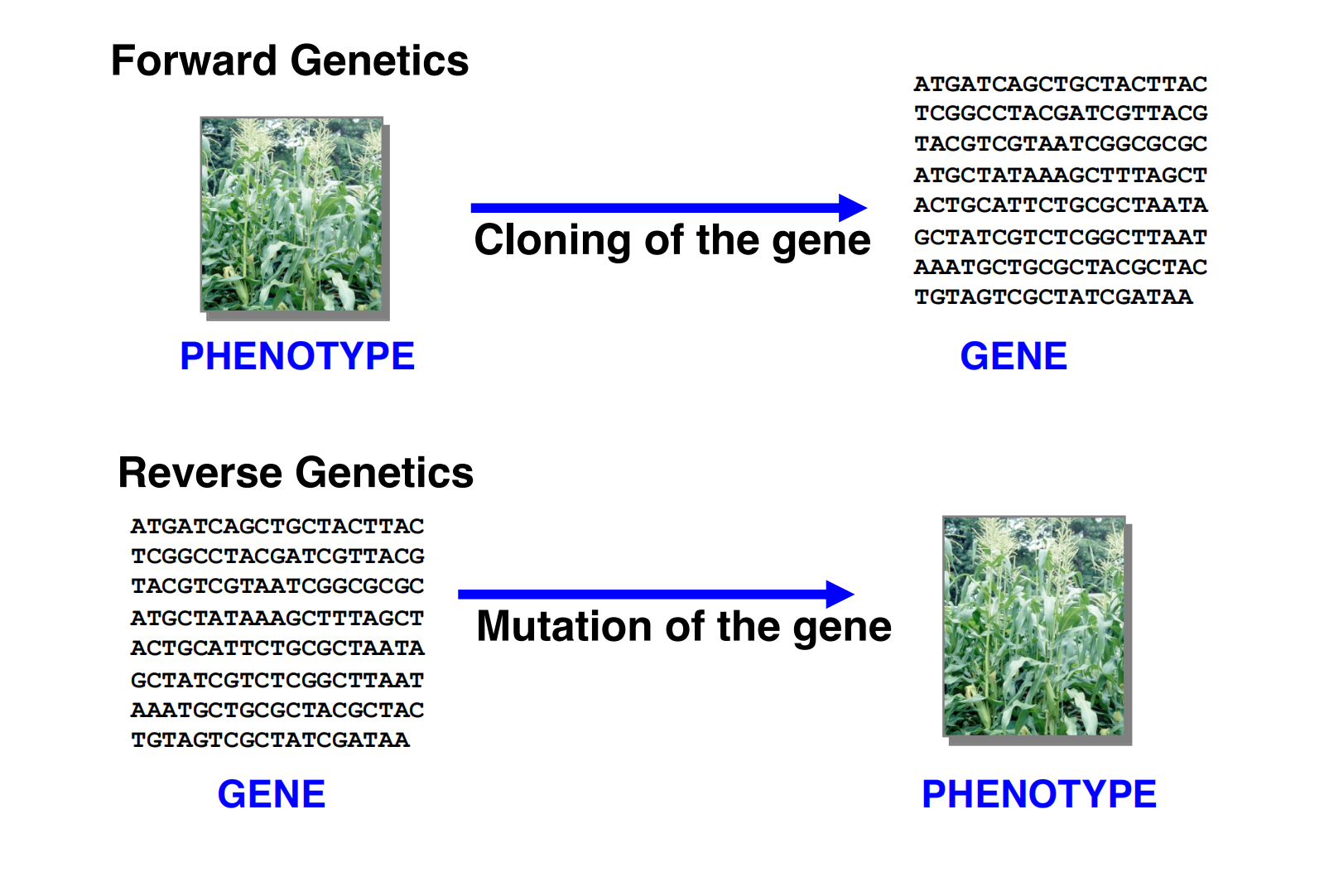
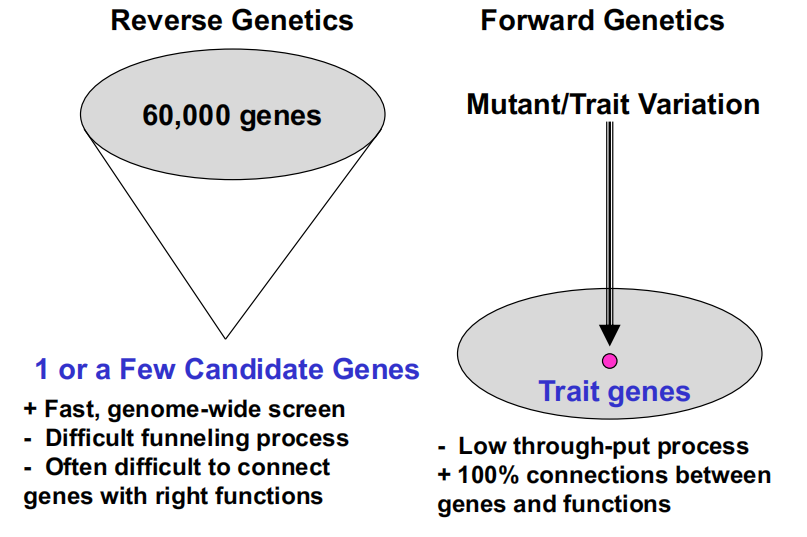
Forward genetics is a phenotype-to-genotype approach
- Connects gene functions to gene sequences
- Forward genetics requires no knowledge of gene products
- Genetic variation or mutations are the starting point
- Mutations induced through mutagenesis
Mutagens most commonly used in plants:
- Fast neutron / gamma rays – large deletions
- EMS (ethyl methanesulfonate) – point mutations
- T-DNA – single insertions, slow to generate
- Transposons – more insertions, easy to generate
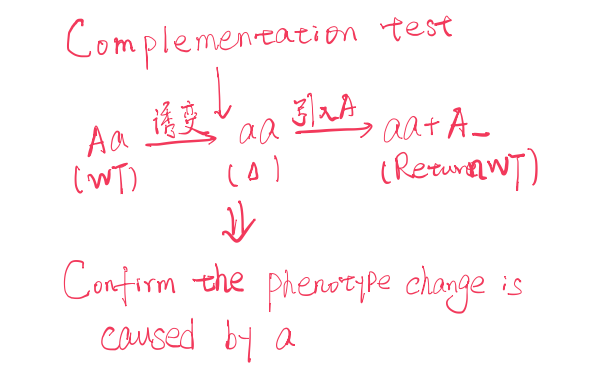
1. Complementation Test
Methods of Forward Genetics
(How can you find mutated genes that cause phenotypes?)
1. Walking – Map-based / positional cloning
(1) Introduction
Look for genetic regions associated with mutations, or exclude chromosomal segments that contain no mutations
- Need for a mapping population
- DNA markers
- Genomic DNA clones (large size)
Map-based Cloning
Map-based cloning is an exercise of converting a genetic map to a physical map
Linkage analysis is the basis of map-based gene cloning
Recombination is the basis of linkage analysis.
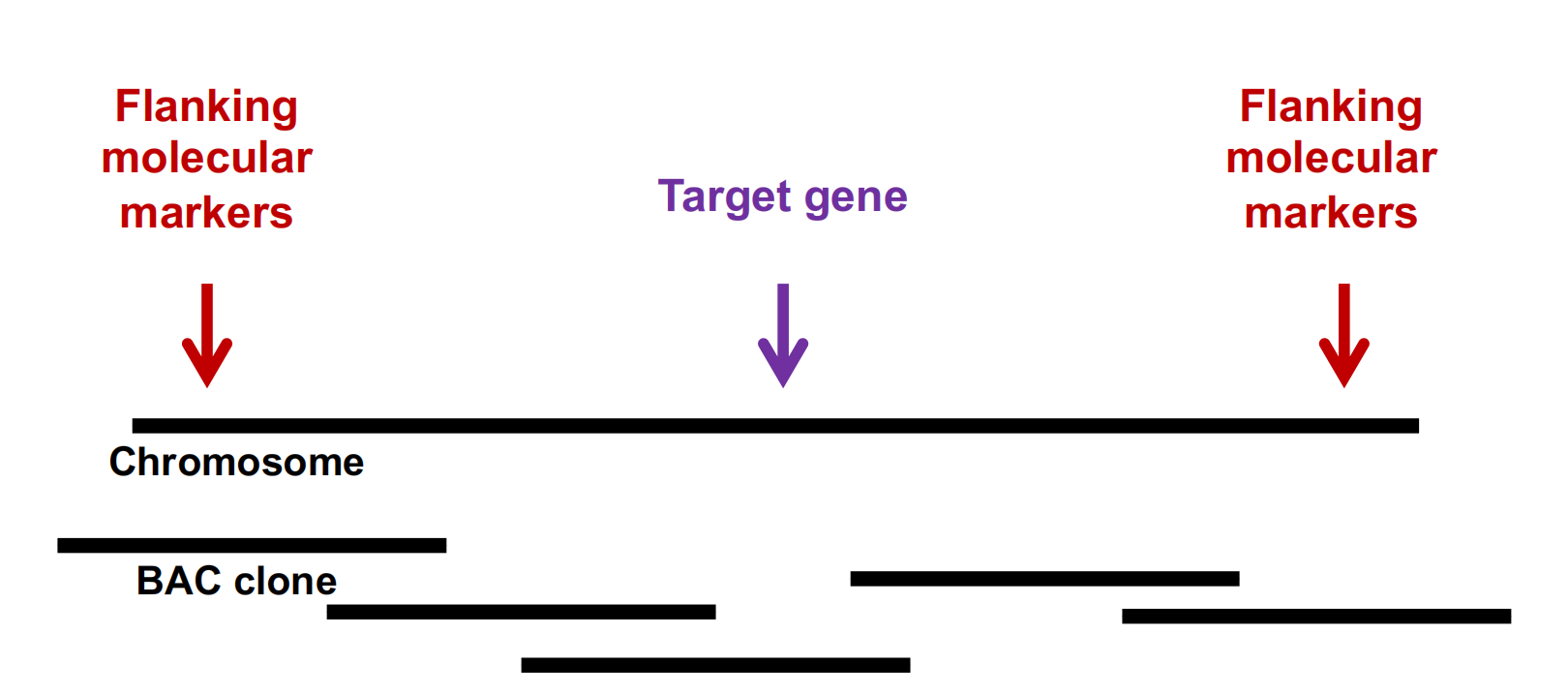
Use genetics to map trait to an interval to find tightly linked markers.
利用遗传学将性状映射到一个区间,以找到紧密相连的标记。
Use marker to identify BAC clone.
BAC(Bacterial Artificial Chromosome,细菌人工染色体)
Use other end of BAC to make a new marker, check for recombinants. Find next clone, repeat until gene is flanked by clones. (找到下一个克隆体,重复这个步骤,直到克隆体包围了基因。)Transform clones to complement mutant. Sequence the gene and determine if the function is known. Sequence clone.
(2) Steps
- Making a mapping population and selection of markers close to the mutant locus
- Selection of large clones covering the mutant locus (Optional when genome sequence is available)
- High-resolution mapping of the mutant locus
- Search for a Candidate Gene and Mutations
- Confirmation of cloning by transgenic complementation
Recombination Frequency Is Proportional to Genetic Distance

1% RF = 1 map unit, a centimorgan (cM)
The closer the distance between two loci, the less frequent is the recombination.
两个基因座之间的距离越近,重组的频率就越小。
High resolution mapping is the process of finding markers that demonstrate fewer or no recombination events.
2. Tagging (transposon or even T-DNA tagging)
Look for genes associated with insertions
- Need for insertion sequences
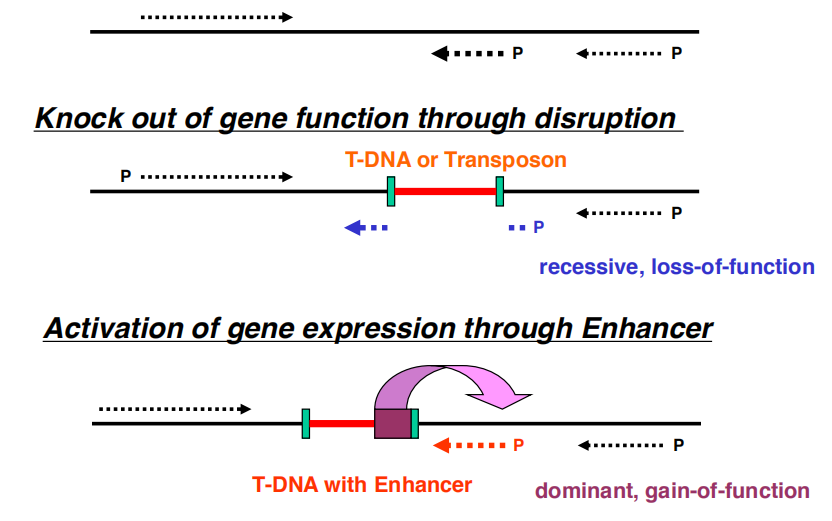
Working Mechanism
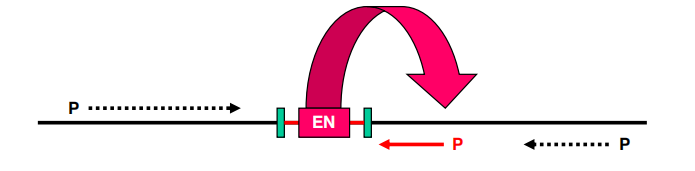
The closest promoter is activated by T-DNA borne enhancer element
EN is not orientation-dependent
- Activation can range over 12 kb
‘One EN-One Gene’ activation
Dominant, Gain-of-Function phenotype
Phenotype can be easily copied in other plants
3. Direct sequencing
Need a good reference genome
- Mutant must be in the same reference genotype
一、Reverse Genetics
Reverse genetics is a genotype-to-phenotype approach that connects gene sequences to gene functions.
Gene sequences are the starting points of reverse genetics (e.g. knowledge of your candidate gene is required)
Characterics of Reverse and Forward Genetics
The Process of Reverse Genetics
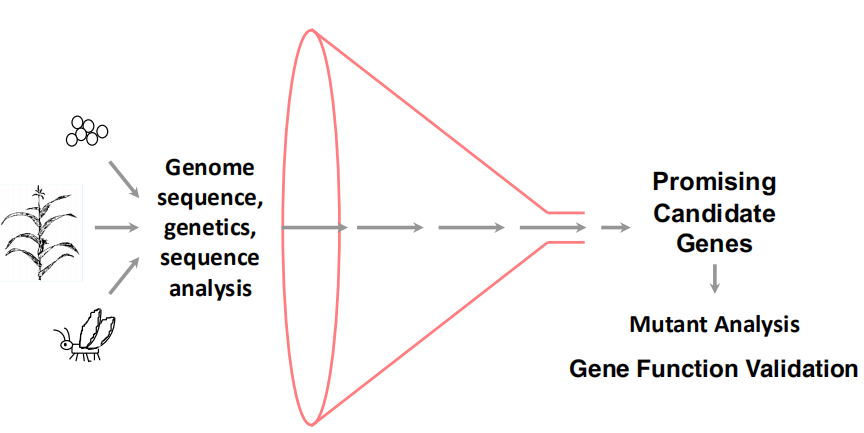
Useful Candidate Data
Expression Pattern – microarray datasets, EST projects, sequence tag libraries, northern analysis, in situ hybridization
Homology – members of an interesting gene family, similarity to genes that may perform a similar function in other organisms
Domain information, such as protein-protein interactions – yeast two-hybrid screen, protein complex isolation/mass spec
Methods of Reverse Genetics: Mutant Approach
How to find mutations that reside in the genes?
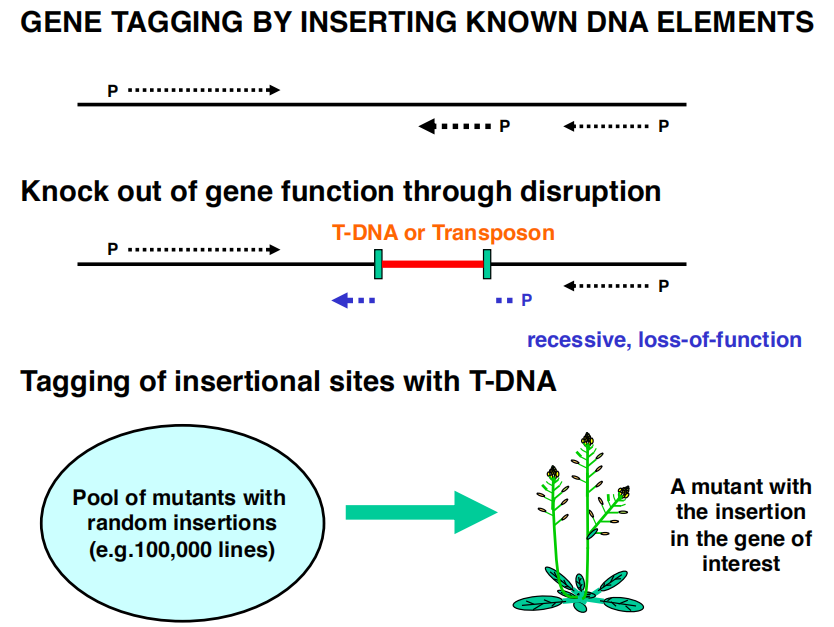
1. Tagging
(T-DNA / Transposon tagging): Look for insertions in gene(s) of interest
- Need an insertion mutant population and DNA from each line
- Gene tagging by inserting known DNA elements
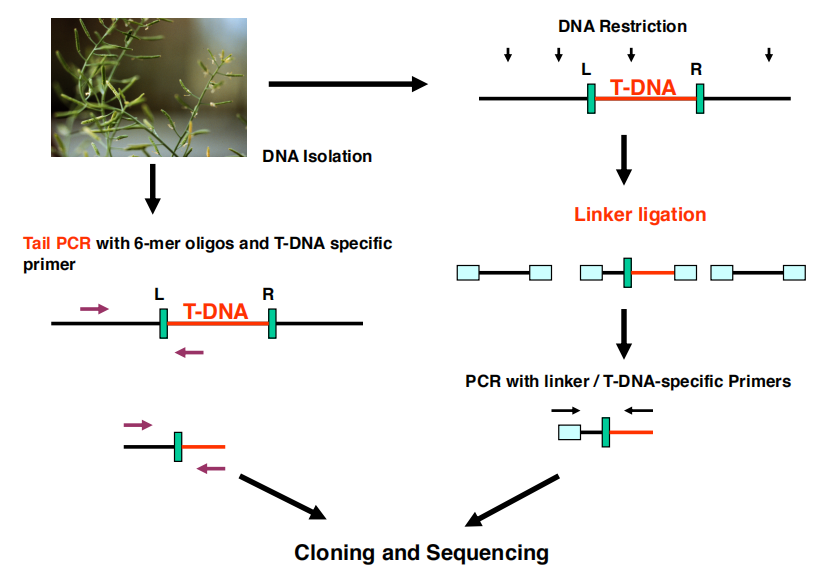
Use PCR to Insert
Amplification of T-DNA insertion sites using two PCR methods
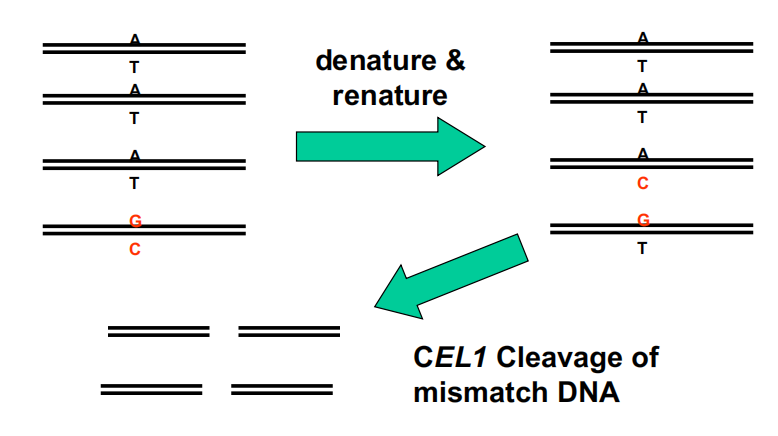
2. Tilling
Look for small changes in gene(s) of interest by analyzing mismatches
- Need a mutant population (EMS, typically)
Tilling technology enables screens to find point mutations in a population