L12 Different Methods for Gene Cloning
General considerations for gene cloning.
- Clone a single gene or making a library ?
- Cloning from DNA or RNA ?
- How to make a cDNA library ?
- How to get full length double-stranded cDNA ?
- End repair of target DNA or RNA
Different systems of cloning
- Restriction-ligation
- Recombinational cloning:Topo Cloning and Gateway cloning
- Seamless Cloning: Gibson Assembly and Golden gate cloning.
一、Clone a single gene or to make a DNA library ?
Possible reason to clone a single gene :
- Expression/purification of a protein
- To study the gene’s function
- To make a mutant
- To complement the phenotype,
- To over-express a gene,
- To make a fusion protein (e.g. To investigate subcellular localization of a protein through fusing with GFP)
Possible reason to make a DNA library:
- To facilitate genome sequencing – think about human genome project, BAC and YAC vectors are developed for this purpose.
- To screening for protein-protein or protein-DNA interaction,
- Yeast two hybrid – Protein-Protein interaction
- Yeast one hybrid – Protein-DNA interaction
- To identify a gene of your interest from the DNA library
二、Clone From DNA or RNA
三、How to make a cDNA Library
Consideration of making cDNA library.
- Capture enough diversity of RNAs.- a few RNA species account for majority of RNA pools.
- Capture the full length of RNA
- RNA integrity
- RNA need to be transformed into dsDNA before it can be cloned as normal DNA library.
- Efficient reverse transcription, second strand synthesis
- Protect the RNA 5’ end
Two typical strategies for preparing cDNA library
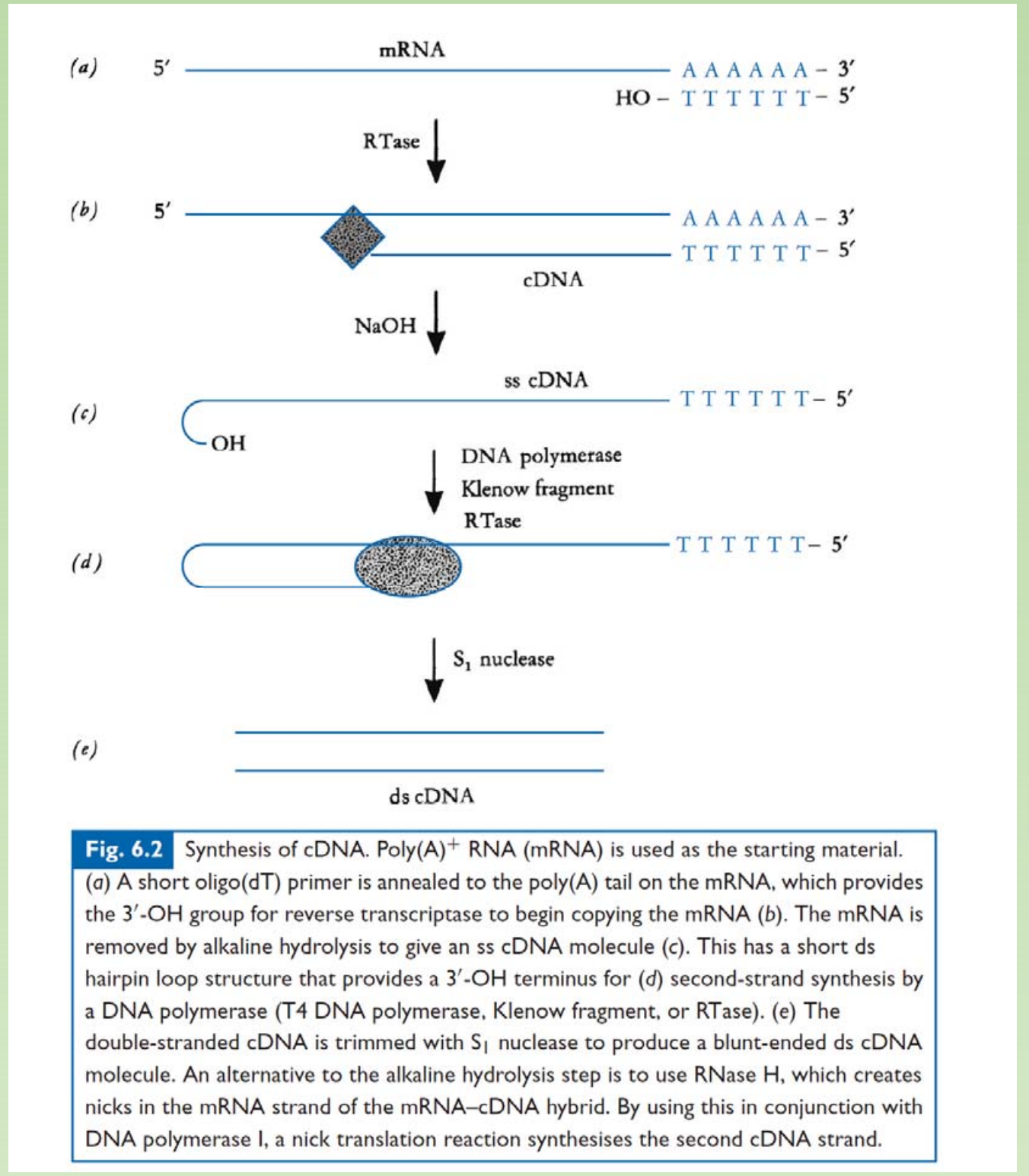
- Reverse Transcription
- Alkaline hydrolysis of RNA template
- A ds harpin loop may form at 3’ end of cDNA.
- A DNA polymerase can use ds harpin loop as primer to synthesize the second strand cDNA.
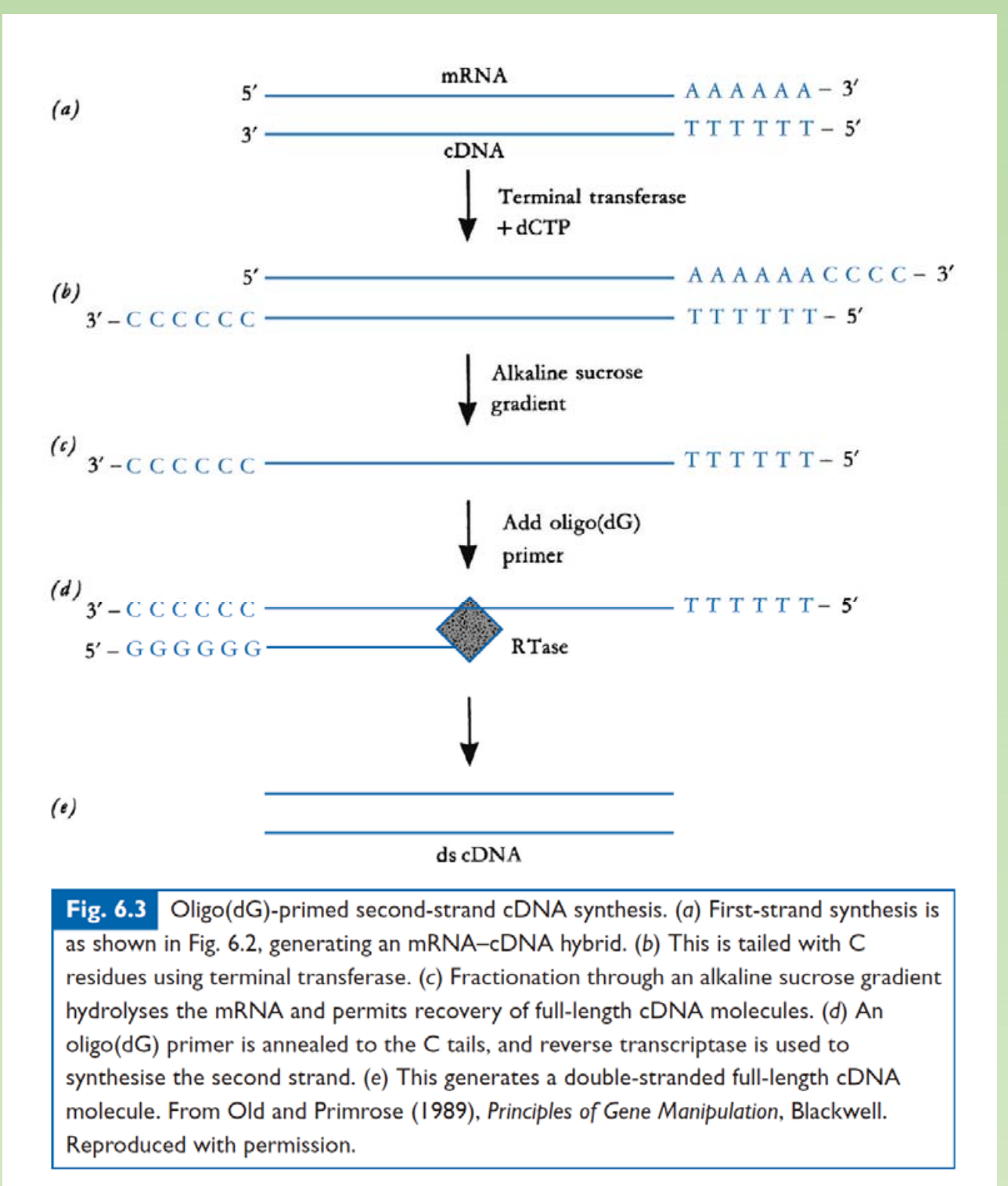
- Reverse Transcription
- TdT + dCTP to do C-tailing on the 3’ end of cDNA.
- Alkaline hydrolysis of RNA template
- Poly G are used as primer to synthesize second strand cDNA.
Disadvantage for these strategies?
- Relatively low efficiency, 5’ end is unprotected. Hard to get full length cDNA.
How to make sure the full-length transcript (cDNA) is enriched ?
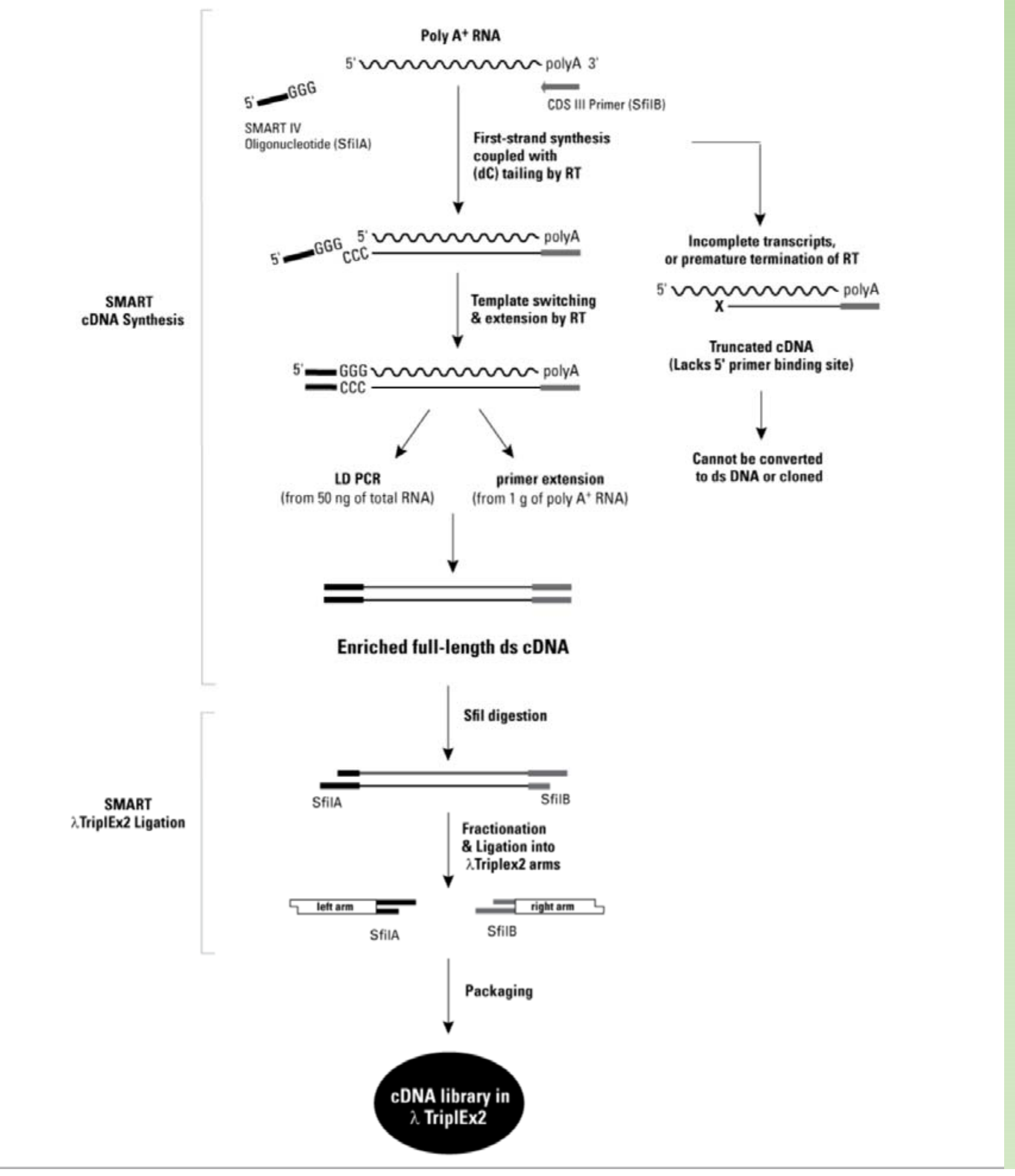
MMLV-RT has a unique property when it finish transcribing the template
- 只有当完成全段转录之后,才会加上三个C,因此确定获得了转录了完成的序列
- MMLV Reverse Transcriptase has terminal transferase activity.
- MMLV add three C deoxyribonucleotide at 3’ end of cDNA (template independent)
- This can be used to anneal with G tailed adaptor.
- Reverse transcriptase switch the template and synthesize the second strand
- This process can largely make sure full length cDNAs are enriched.
- The protocol is very useful in long reads sequencing (PACBIO or Nanopore)
A few screening methods of DNA library
1. Screen the library by dot-blot
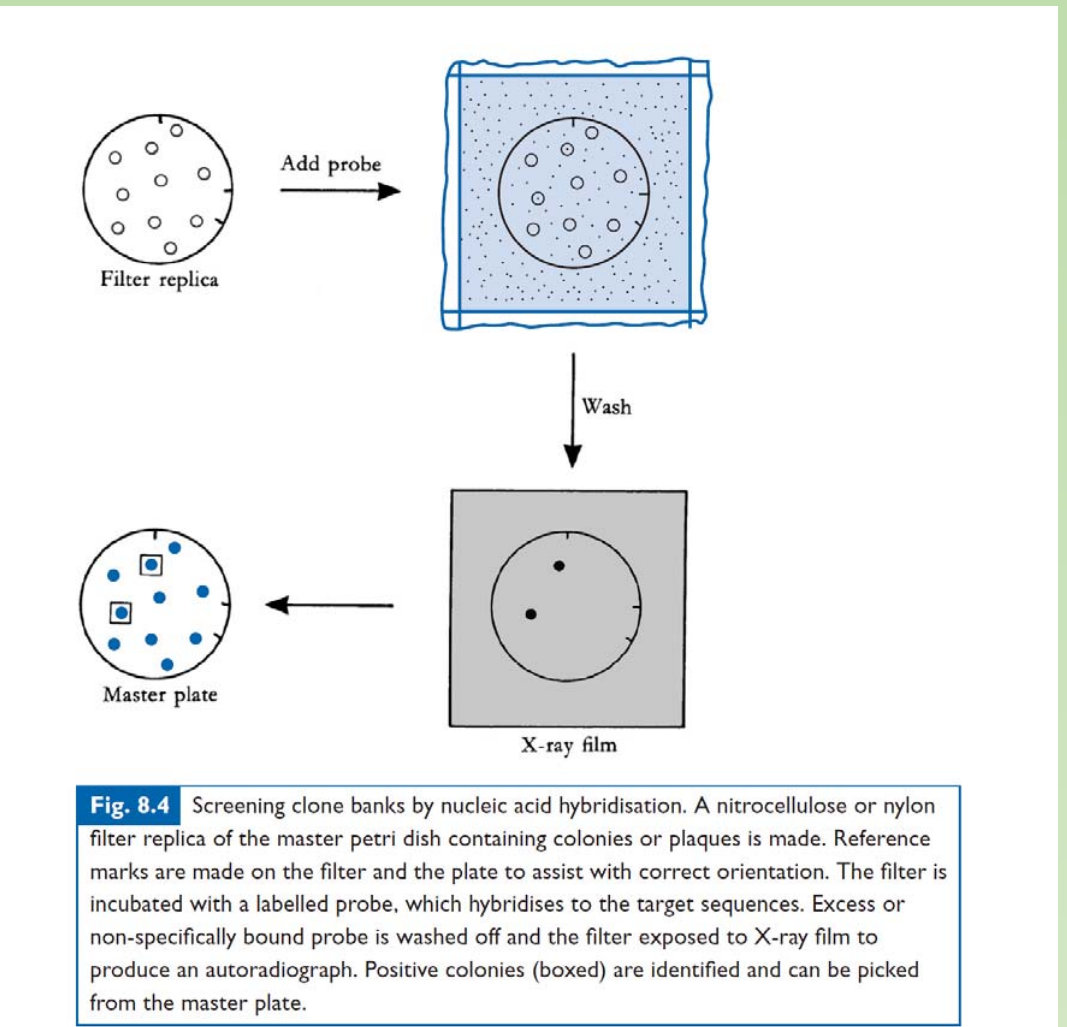
Screen the library by dot-blot
- Making labeled probe of your target
- Filter replica the plate
- Hybridization
- Find the colony or plaque
2. Plus/minus Screening
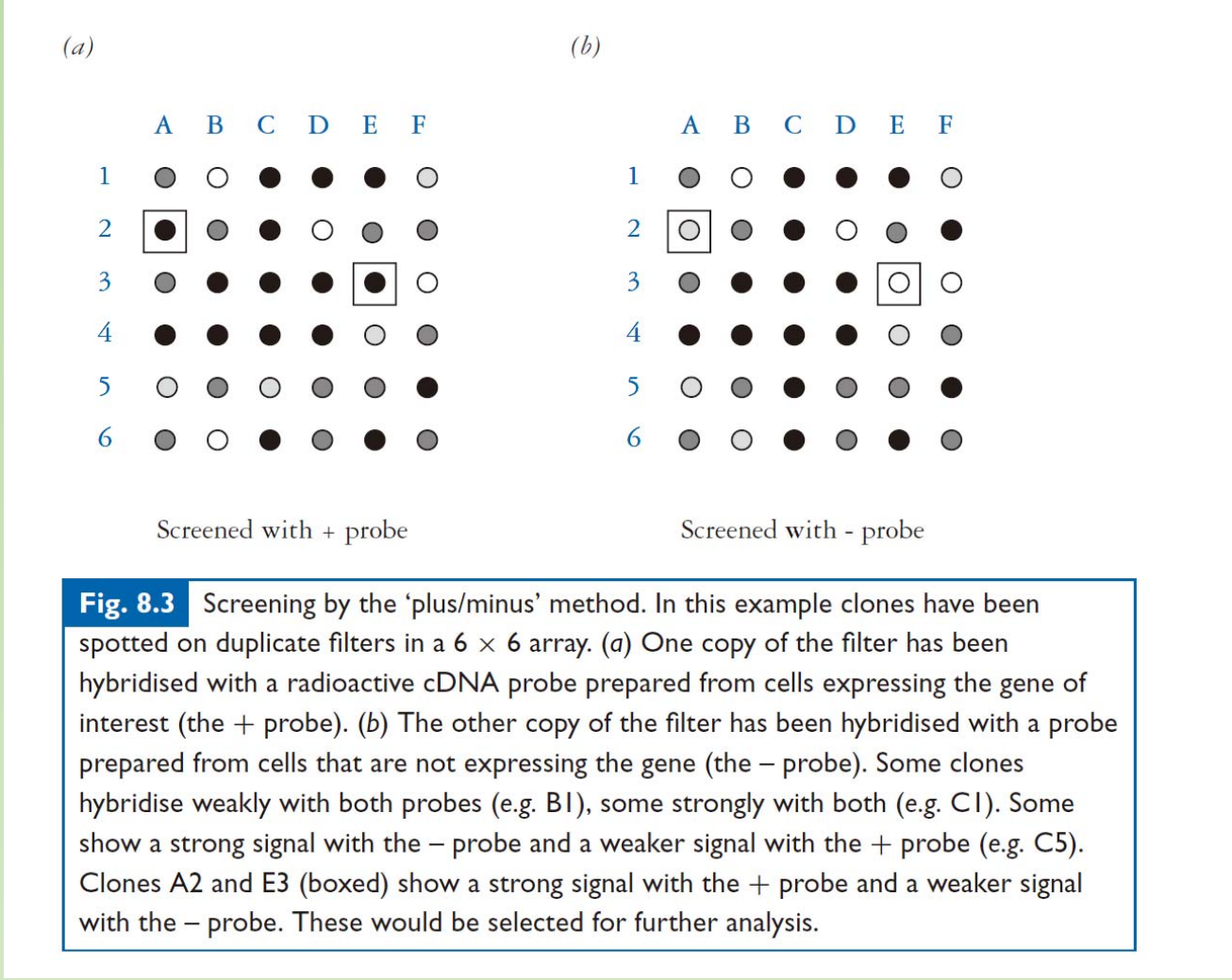
Plus/minus Screening
- To identify gene of interest that only expressed under certain condition
- Assuming you want to identify a gene that only express in condition A.
- to label : put redioactive sub stances during the transcript ion
- Plus probe: labeled cDNA that made from condition A.
- Minus probe: labeled cDNA from condition B.
3. Yeast two-hybrid
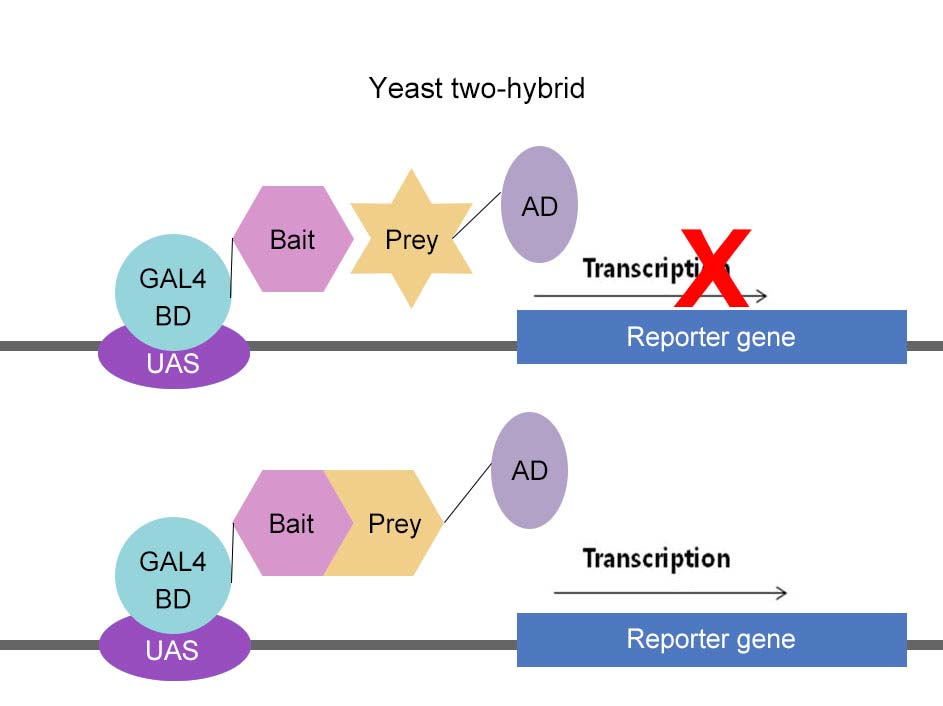
- To identify the interaction partner of the protein of interest
- The connection between GAL4 DNA-binding domain (BD) and GAL4 activation domain (AD) through the interaction between Bait protein and Prey protein activate reporter gene
- cDNA library are often made as AD fusion.
Typical methods of generating the target fragment from DNA or RNA template.
PCR – for individual gene cloning
Mechanic force – e.g. sonication
Enzymatic cut – Restriction Enzyme, Dnase I, MNase, RNase etc.
Direct synthesis
Reverse-transcription (For RNA, the rest is the same as for DNA)
End repair of target DNA or RNA
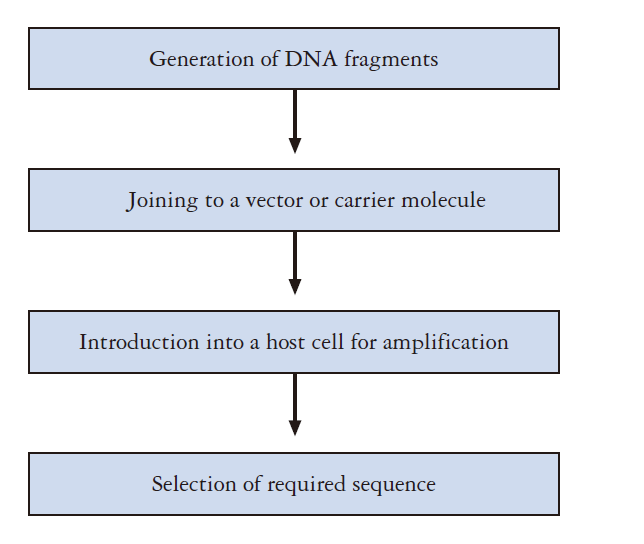
How to put the DNA fragment into vector ?
- most reliable/classical way is ligation.
There are few steps may be needed before we do the ligations
End repair Add or Remove OH or PO4 group
- T4 PNK and Alkaline phosphatase
- MNase cut generate terminal 3’-phosphates
- To prevent self-ligation
Blunting – remove 3’ overhang and filling 5’ over hang (T4 DNA Pol)
Homopolymer Tailing – TdT (Terminal Deoxynucleotidyl Transferase ).
Linker/adaptor addition to facilitate ligation with vector or prevent the cutting of fragment(mostly used in cDNA library).s
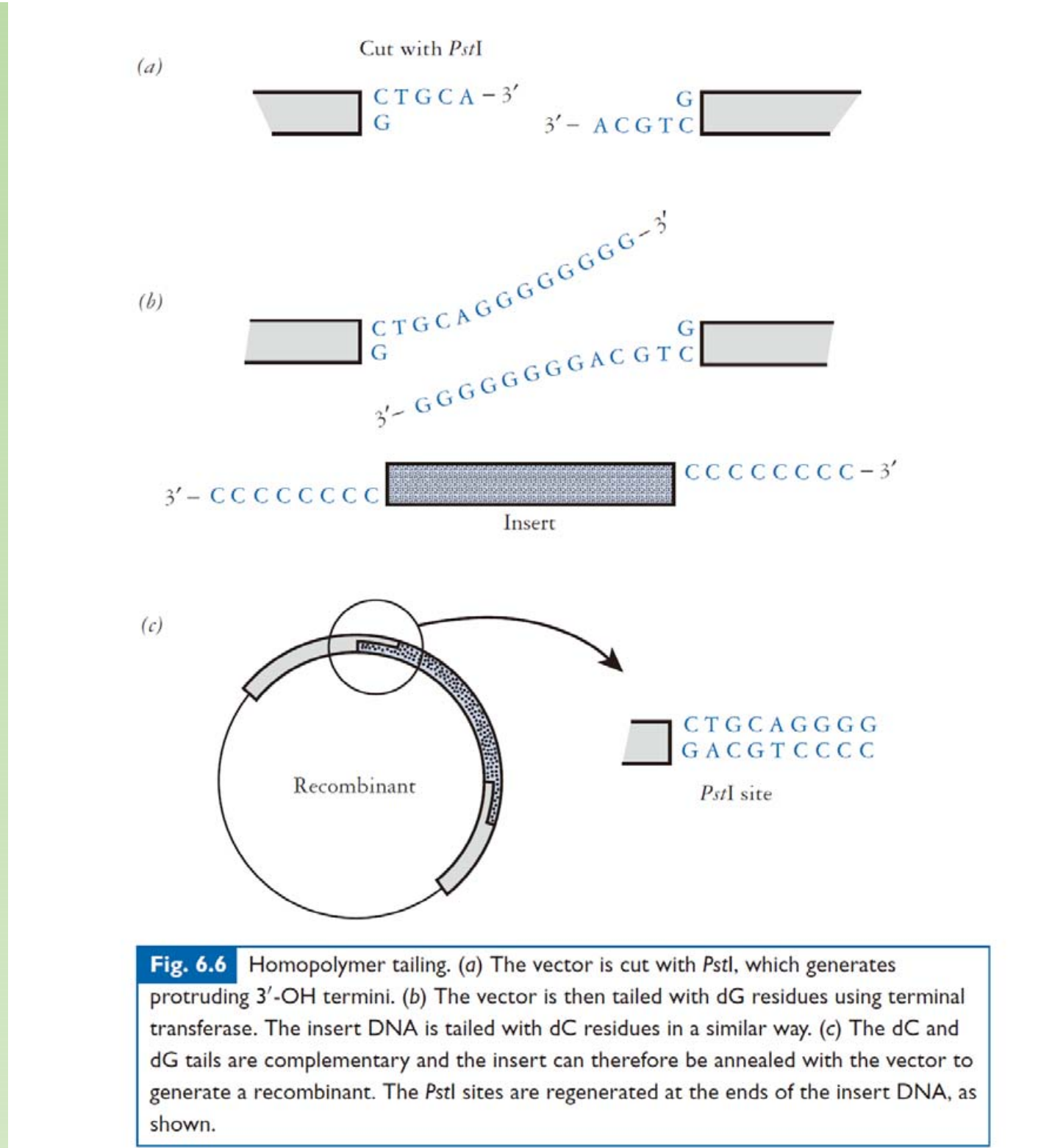
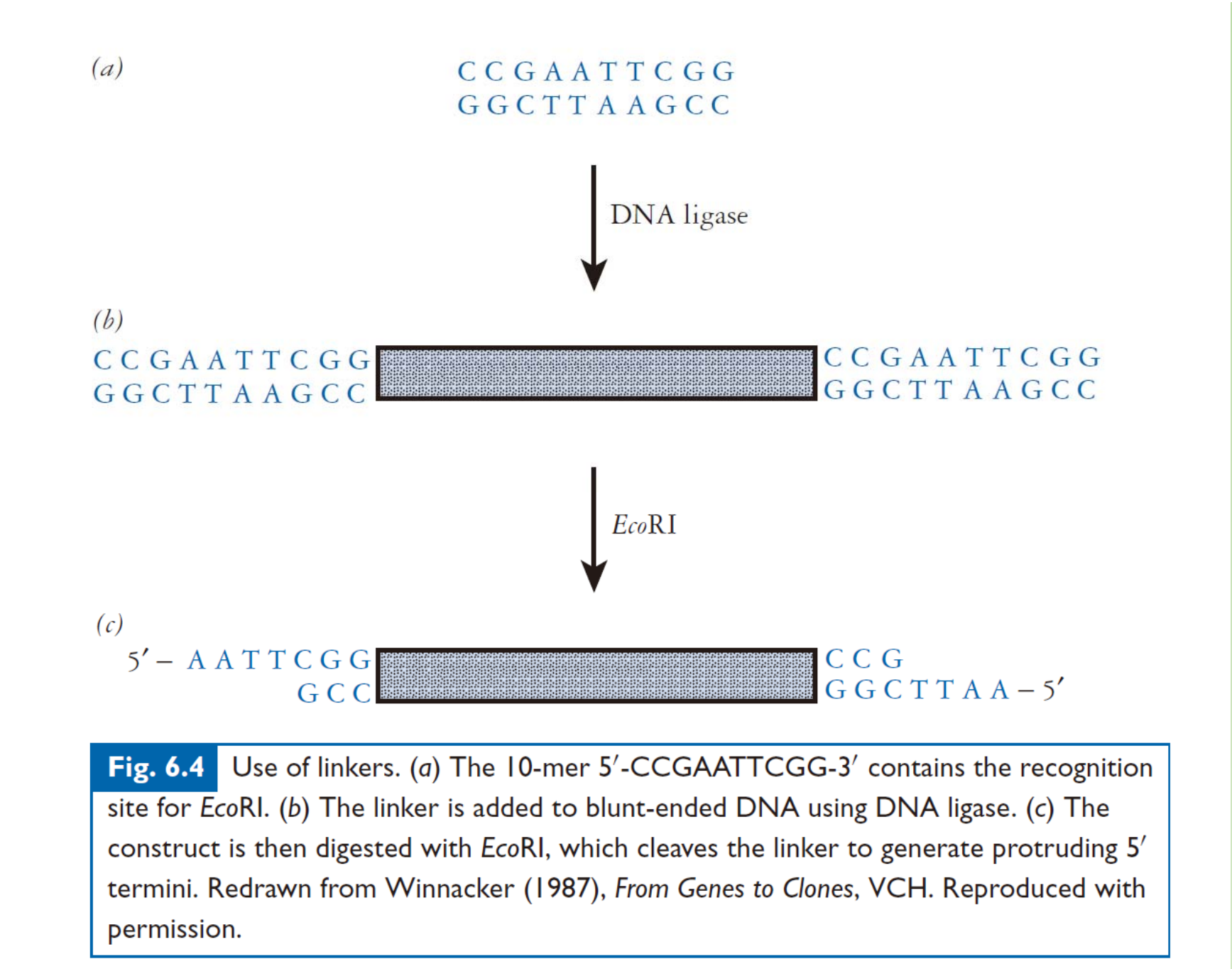
四、Different systems of cloning
Recombinational cloning
These systems use a site-specific recombinase. DNA is recombined with each other through specific sequences that is added at both end of the fragment or vector. Typical system now available is Gateway cloning system.
This recombination reactions are facilitated by the recombination of attachment sites from the phage (attP) and the bacteria (attB).
The recombined site called attL (left) and att R(right).
att B1 fragment attB2 + attP1 -ccdB-att P2 = att L1 fragment att L2 + att R1 ccdB + att R2
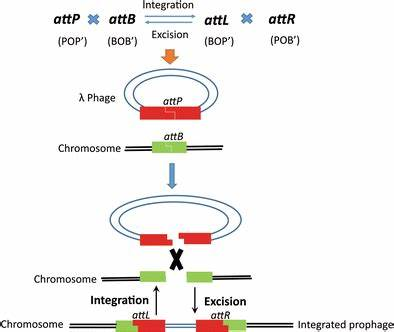
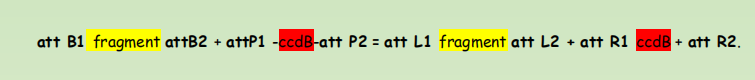
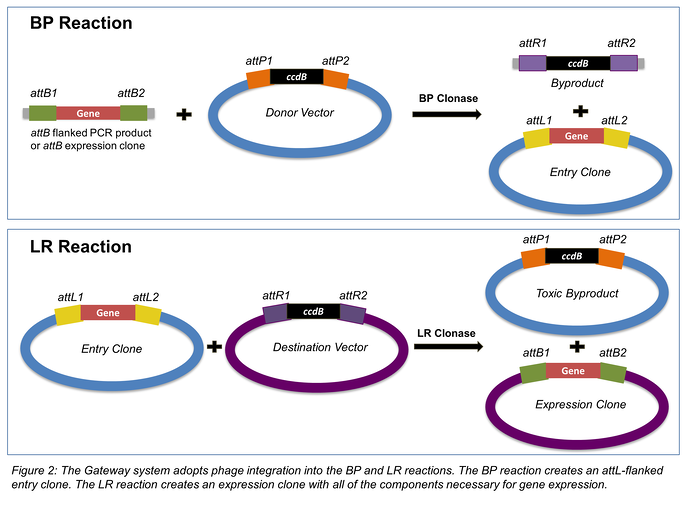
- Donor vector
- Entry vector
- Destination vector
- BP clonase and LR clonase
Pros and Cons of Gateway cloning system
Advantage:
- LR and BP reaction are highly accurate.
- Many different choice of destination vector available.
Disadvantage:
- Very expensive
- Depends on the availability of destination vector.
Topo Cloning
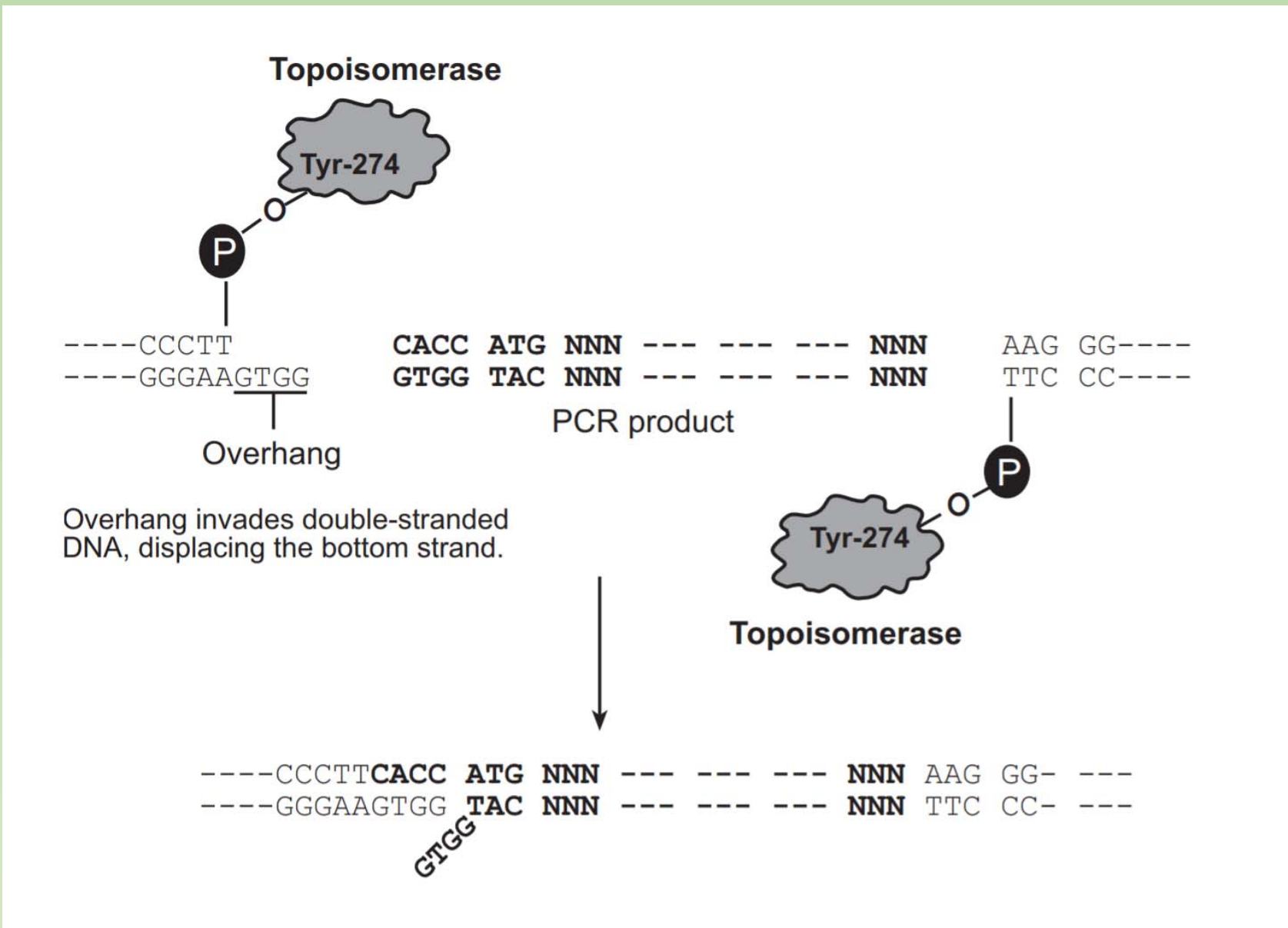
- The TOPO technique specifically uses Vaccinia virus-isolated topoisomerase I.
- Apart from topoisomerase activity, this enzyme recognizes the DNA sequence 5´(C/T)CCTT-3‘ and digests double stranded DNA at this sequence.
- The energy from this breakage is stored as a covalent bond between the cleaved 3′ DNA strand and a tyrosyl residue (Tyr 274) of topoisomerase I.
- If a 5′ hydroxyl group from a different DNA strand comes along, it can attack this covalent bond thus joining the two DNA strands and releasing topoisomerase.
Advantage:
- No post PCR procedure is needed.
- Can be directional
Disadvantage:
- Expensive
- Low efficiency when dealing with fragment >3 kb.
Seamless Cloning: Gibson Assembly

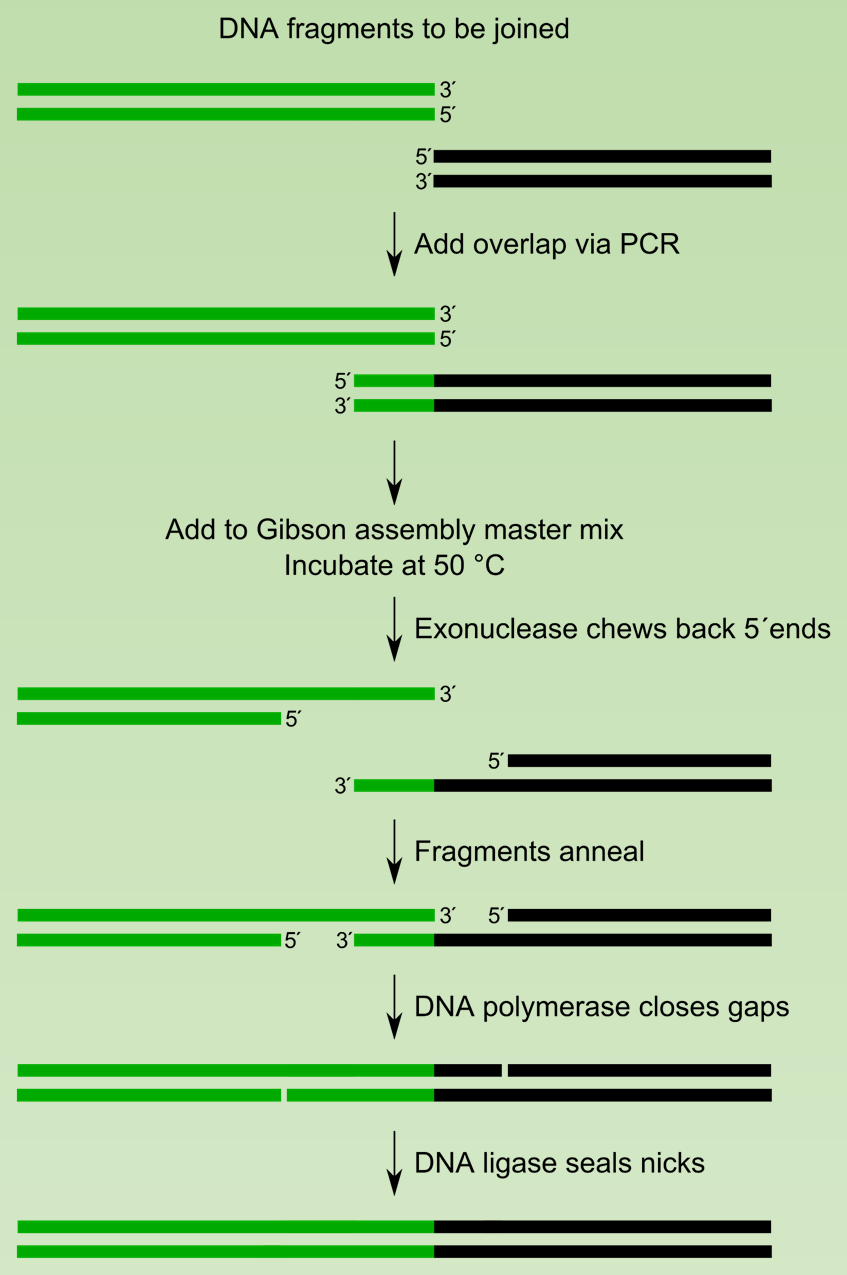
- Exonuclease degrade 5’ to 3’ and create 3’ overhang
- Overhang between fragment form double stranded region.
- DNA polymerase fills the gap created by exonuclease.
- DNA ligase seal the nick.
Drawbacks:
- The original method is inefficient in cloning fragment shorter than 50bp, due to the activity of exonuclease. Cloning efficiency can be reduced if there is complicated secondary structure in the overhang region
3’ to 5’ Exonuclease can also be used in Seamless cloning
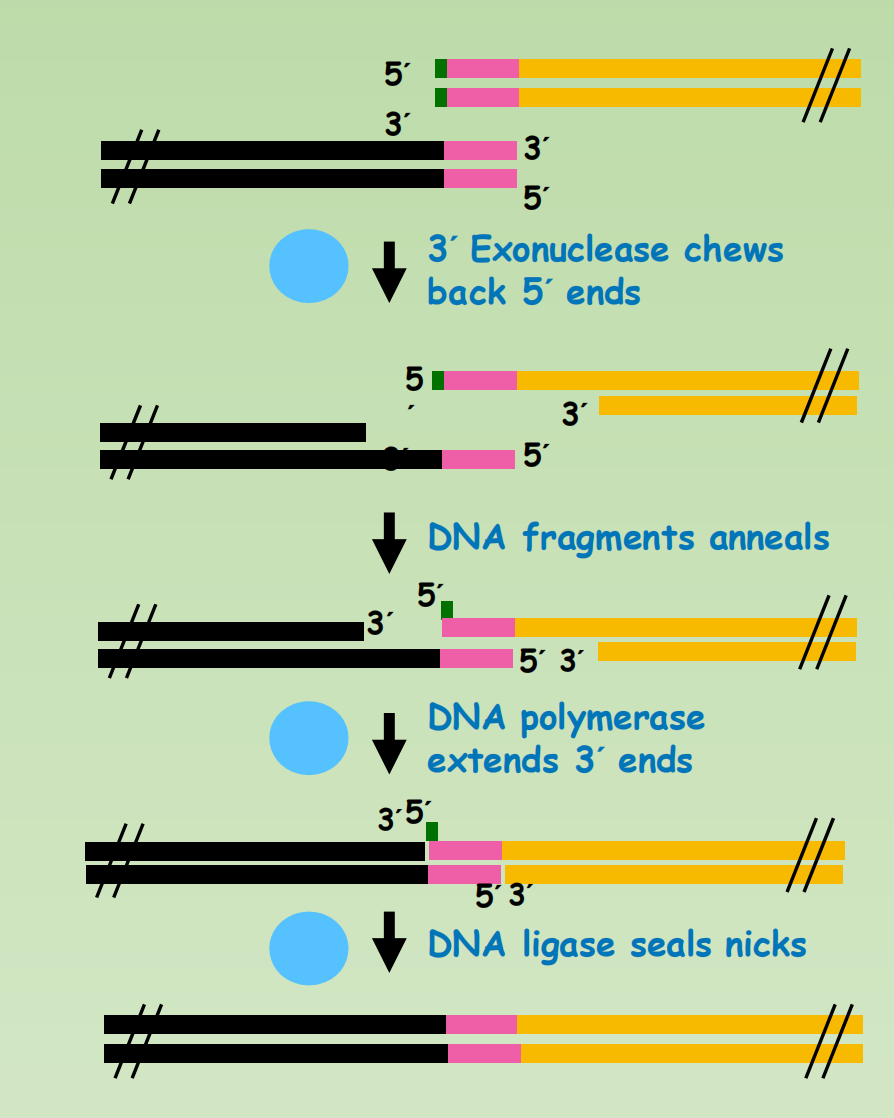
Exonuclease: The exonuclease activity defines which strand (if any) gets digested to expose complementary overhangs:
- 5’ exonuclease (Gibson & GeneArt® Seamless Cloning): The enzyme chews back bases from the 5’ end to expose complementary overhangs. GeneArt® Seamless Cloning recommends a 15 bp overhang, while to perform a Gibson assembly a longer overhang of 25 to 40 bp is used in many protocols.
- 3’ exonuclease (SLIC & In-Fusion® Cloning): The enzyme chews back bases from the 3’ end to expose complementary overhangs. A 15 bp overhang is recommended for In-Fusion® Cloning, while for SLIC a 25 bp overhang is recommended.
Do we really need DNA polymerase and DNA ligase ?
- Seems not!
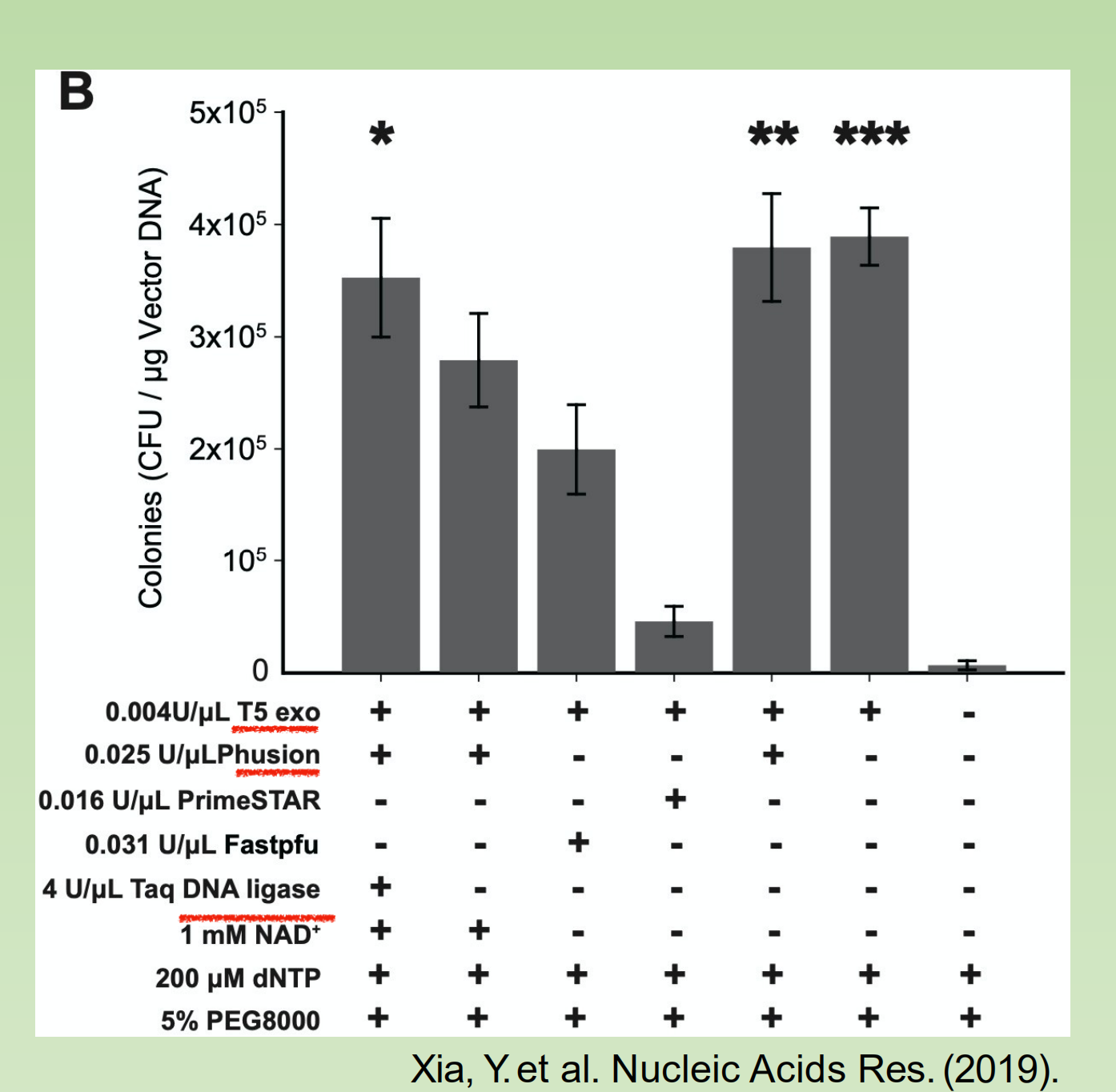
TEDA
- (T5 exonuclease DNA assembly)
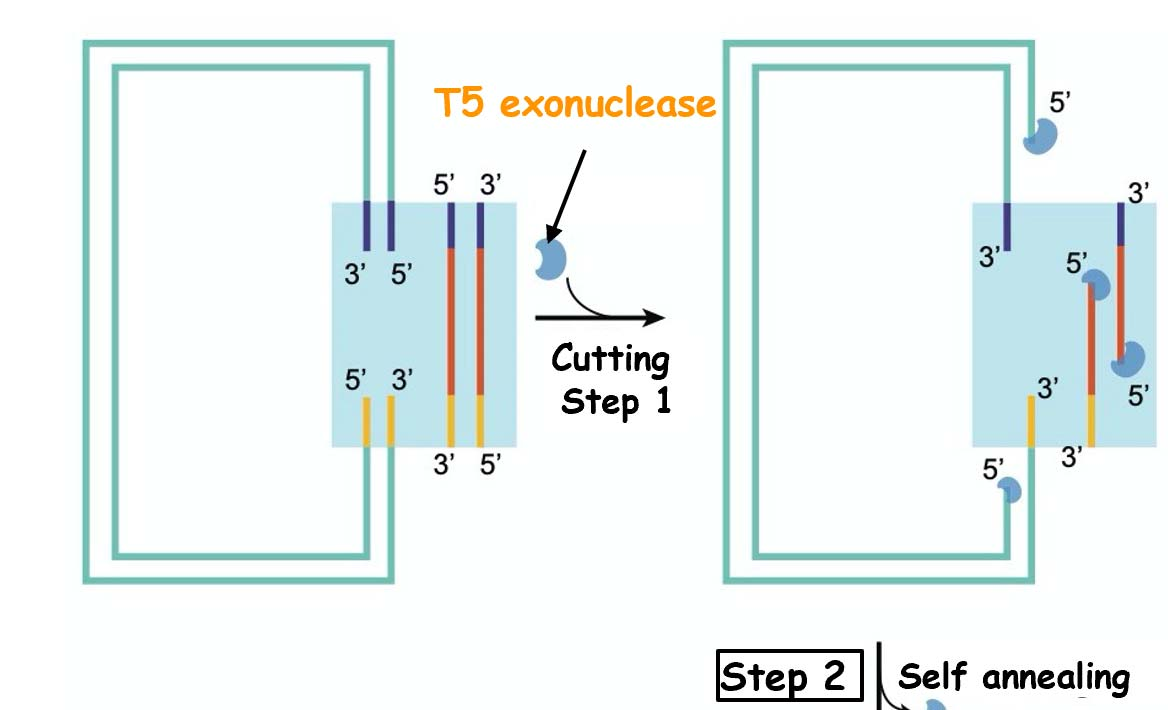
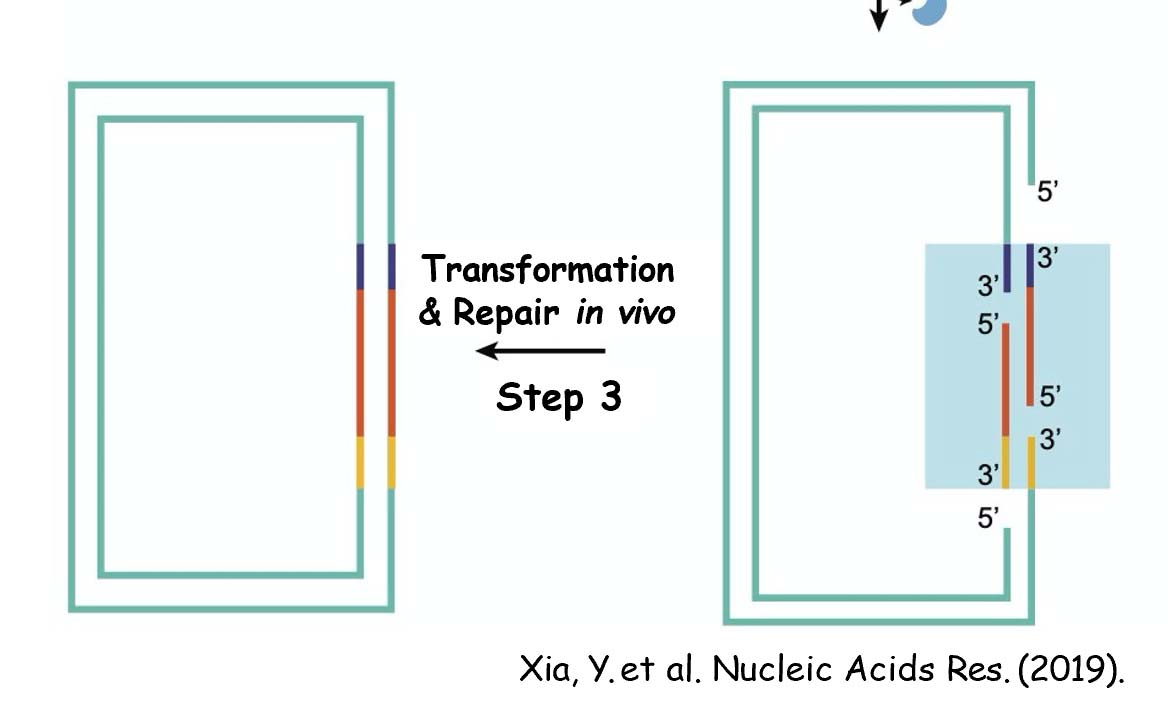
T5 Exonuclease degrades DNA in the 5´ to 3´ direction.
3´ direction. T5 Exonuclease is able to initiate nucleotide removal from the 5´ termini or at gaps and nicks of linear or circular dsDNA. However, the enzyme does not degrade supercoiled dsDNA

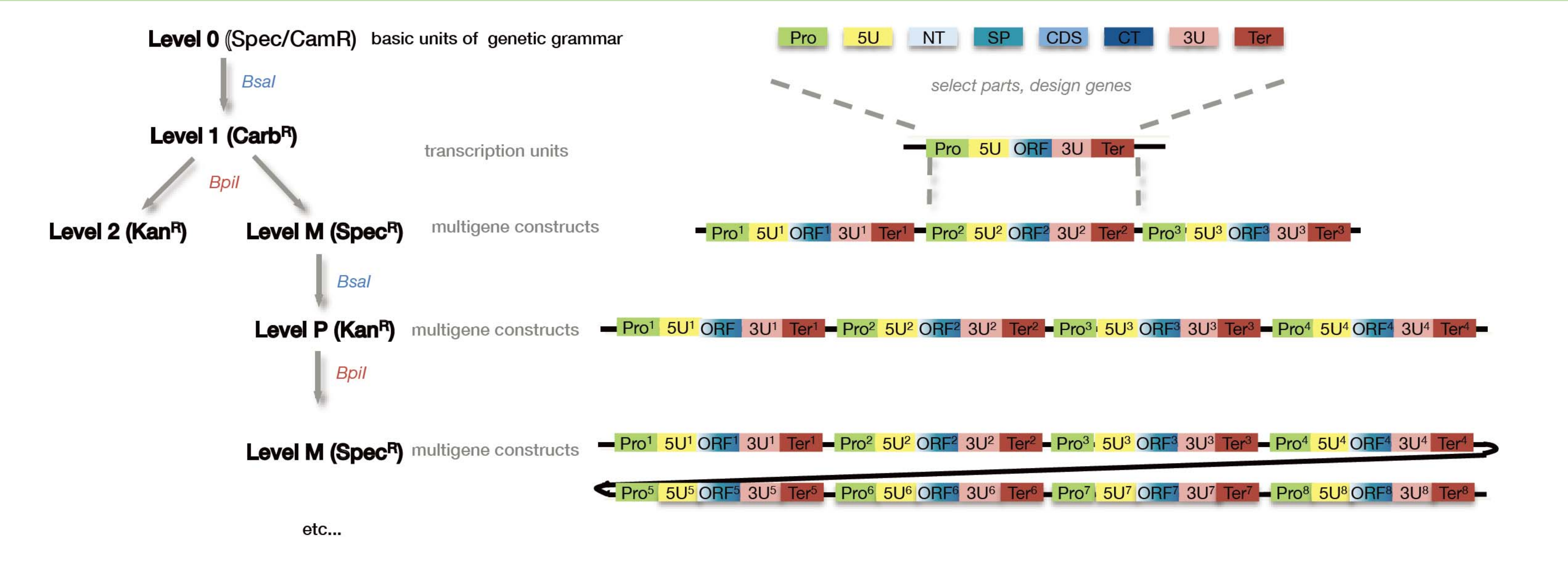
Seamless Cloning: Golden gate cloning
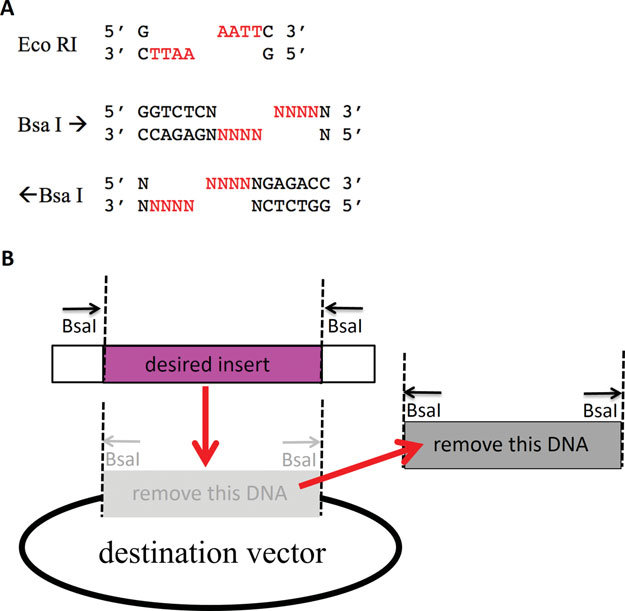
Type II S restriction enzyme:
- Type IIS restriction enzymes recognize asymmetric DNA sequences and cleave outside of their recognition sequence
BSA I and BPI I
- The overhang sequence created is not dictated by the REase, and therefore no scar sequence is introduced
- The fragment-specific sequence of the overhangs allows orderly assembly of multiple fragments simultaneously
- The restriction site is eliminated from the ligated product, so digestion and ligation can be carried out simultaneously
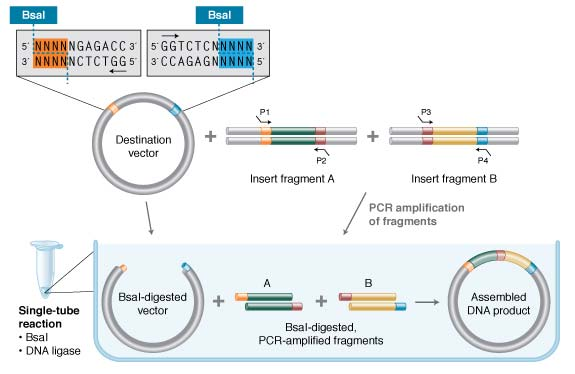
- P1, P2, P3, P4 are overhang that generated by BsaI cut, which can be designed as you want.
- The generated compatible overhang can be ligated by DNA ligase
- Once ligated, it can not be cleaved again by BsaI.
- Cycles of BsaI digestion and DNA ligation are often performed.
- By this end of several cycle, there will be quite a lot of ligated products
1. Typical protocol for Golden Gate Cloning
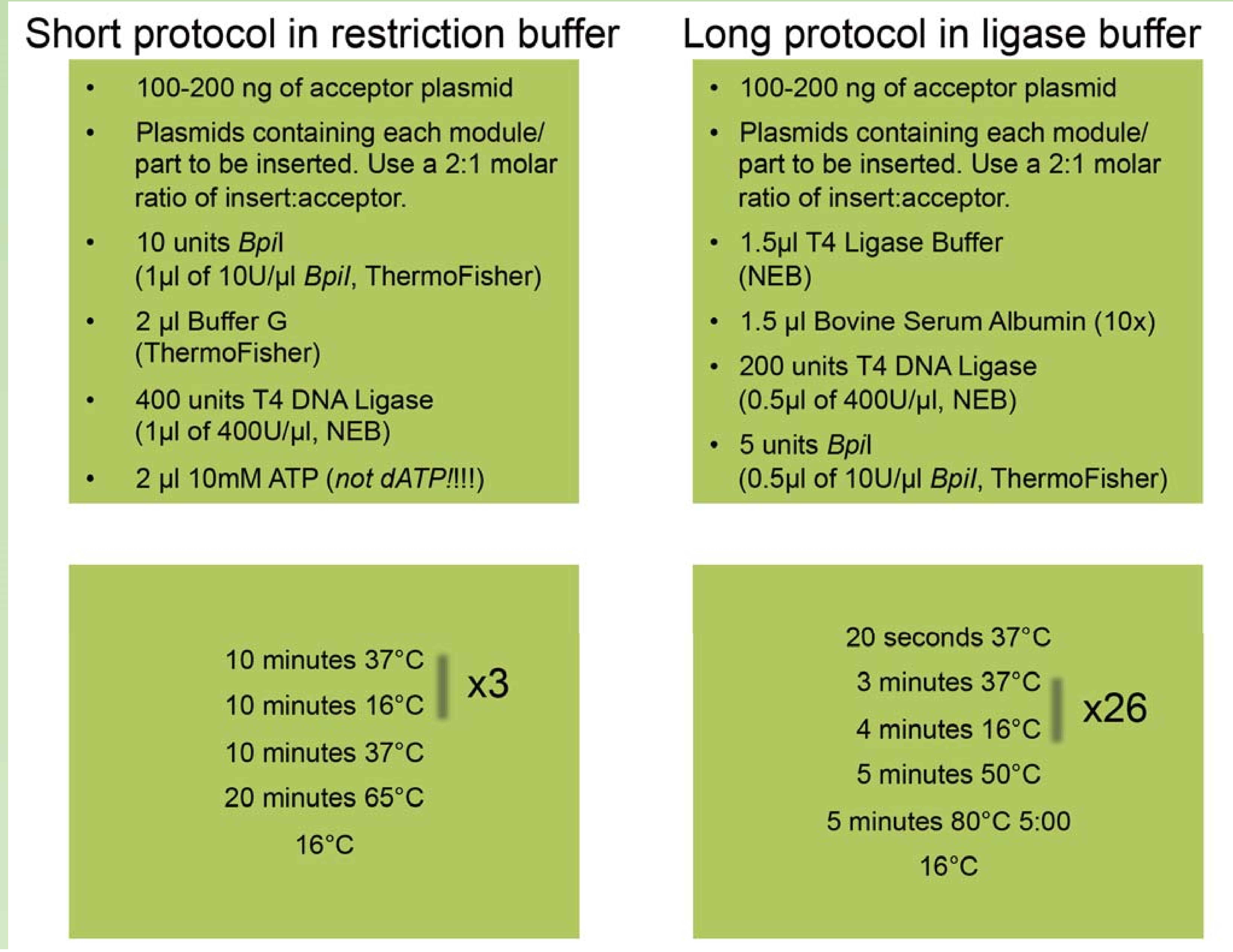
- why the cycles of restriction cut and ligation is required ?
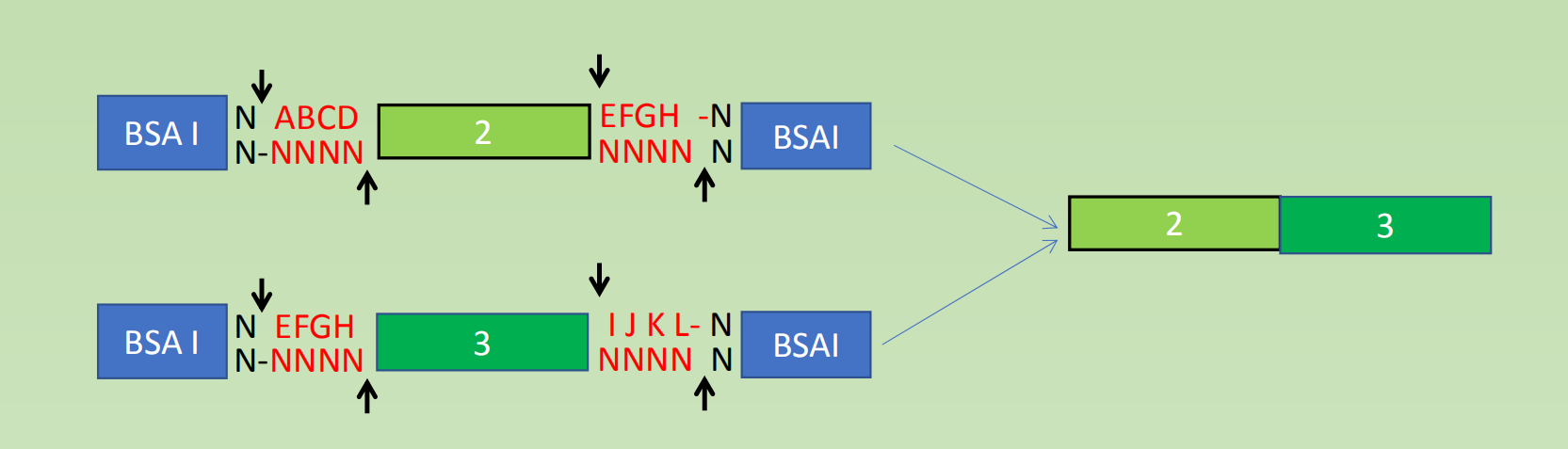
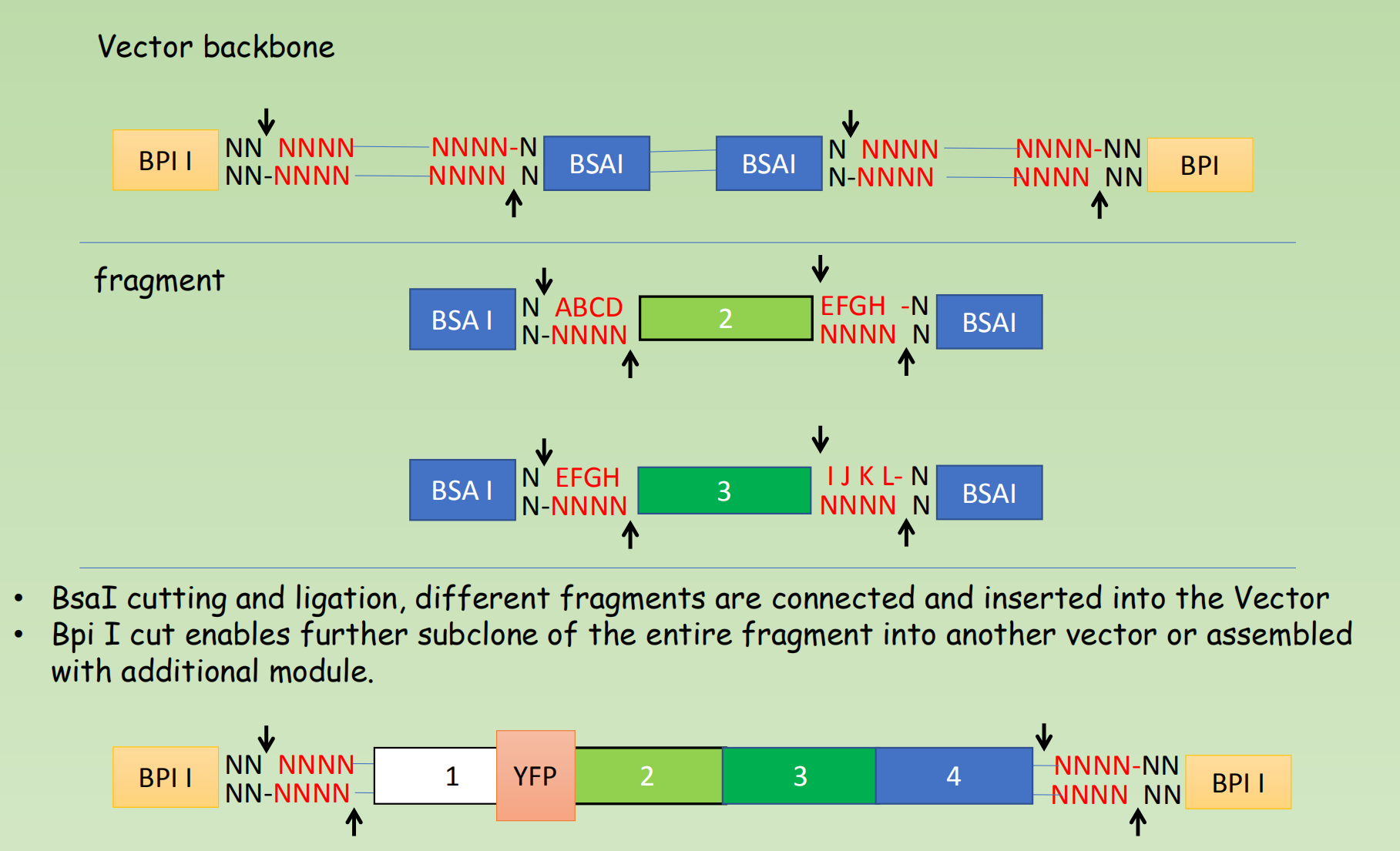
- BsaI cutting and ligation, different fragments are connected and inserted into the Vector
- Bpi I cut enables further subclone of the entire fragment into another vector or assembled with additional module
2. Advantages of the Golden Gate cloning
The Golden Gate ligation process is close to 100% efficient thanks to re-digestion mechanisms. Also, re-ligation is prevented, because cleaving outside of restriction enzymes sites removes them from the product. Good to make an array of repeat sequences, TALEN.
3. Disadvantage
Much more sequence dependent than other method, there should be no BsaI site within the sequence. This may be less of a problem when you directly synthesize the fragment as you can mutate the sequence.