L04 Electron Transport, Oxidative Phosphorylation, and Oxygen Metabolism
一、The Mitochondrion: Scene of the Action
The average adult human synthesizes ATP at a rate of nearly 10^21^ molecules per second, equivalent to producing his or her own weight in ATP every day.
Glycolysis and the citric acid cycle by themselves generate relatively little ATP directly.
However, under aerobic conditions, six dehydrogenation steps:
- One in glycolysis
- One in pyruvate dehydrogenase
- Four in the citric acid cycle
Collectively reduce 10 moles of NAD+ to NADH and 2 moles of FAD to FADH2 per mole of glucose.
Reoxidation of these reduced electron carriers in the process termed cellular respiration generates most of the energy needed for ATP synthesis.
Oxidation of 1 mole of NADH (FADH2) by the respiratory chain provides sufficient energy for synthesis of 2.5 ( 1.5) moles of ATP from ADP.
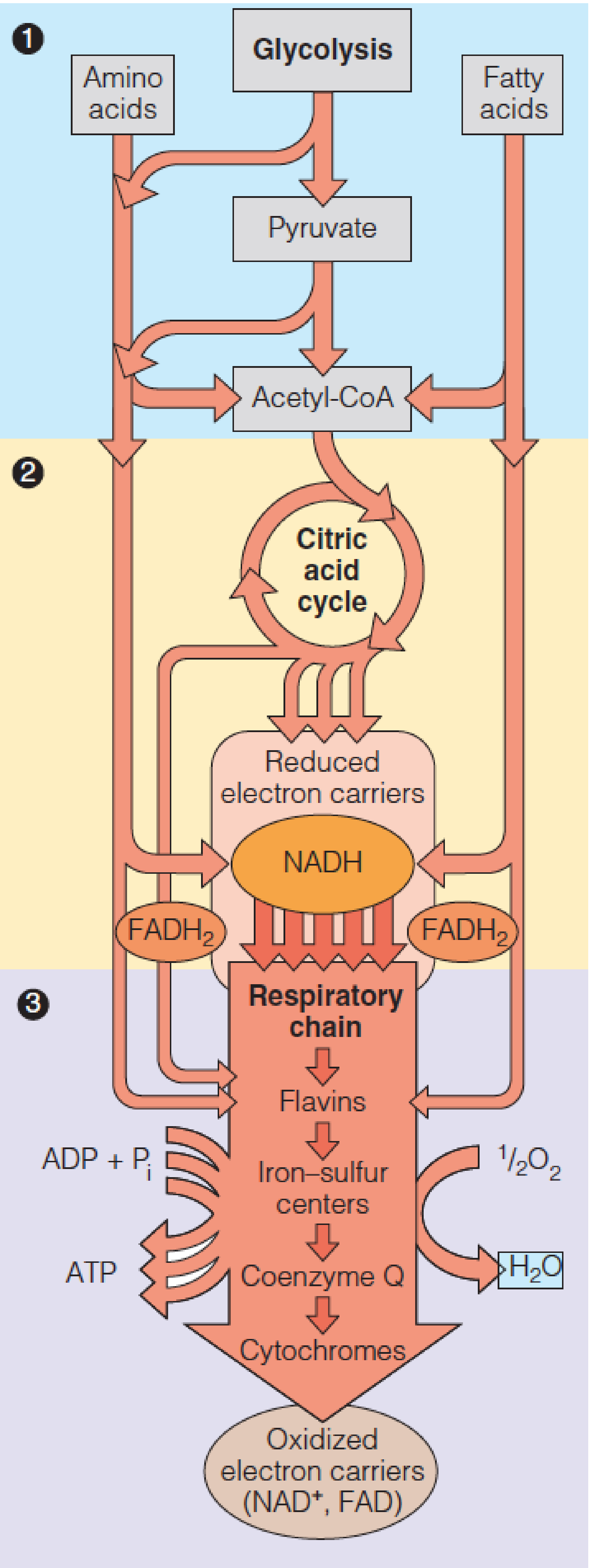
In eukaryotic cells NADH and FADH2 are reoxidized by electron transport proteins bound to the inner mitochondrial membrane.
A series of linked oxidation and reduction reactions occurs, with electrons being passed along a series of electron carriers—the electron transport chain.
The final step is reduction of O2 to water.
The overall electron transport sequence is quite exergonic.
One pair of reducing equivalents, generated from 1 mole of NADH, suffices to drive the synthesis of between 2 and 3 moles of ATP from ADP and by a process termed oxidative phosphorylation.
(32 x 30.5)/2870 = 34% under standard conditions
ATP hydrolysis is worth 50-60 kJ/mol in vivo, 60-70%
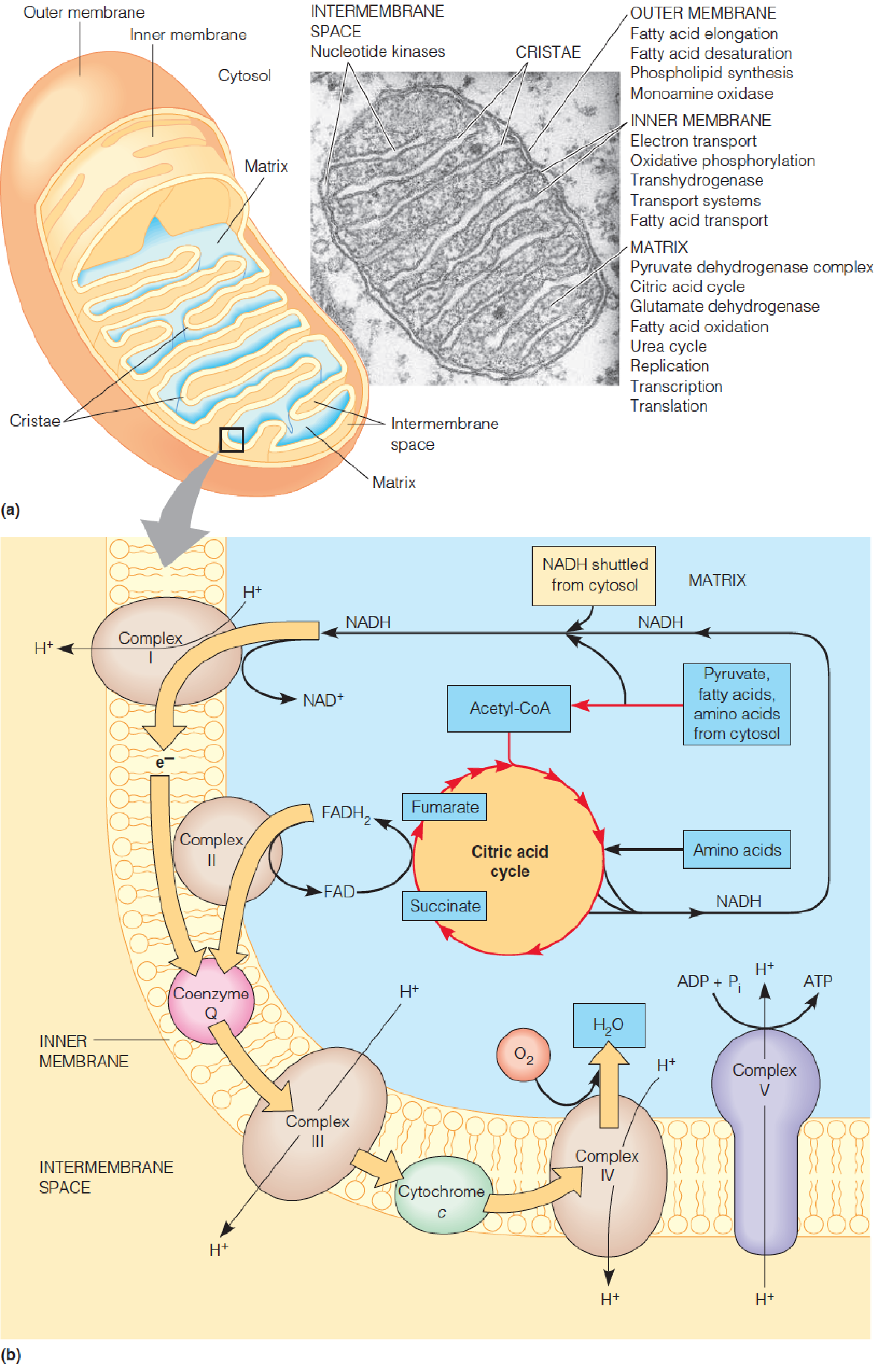
Localization of respiratory processes in the mitochondrion
A mitochondrion from a pancreatic cell.
Overview of oxidative phosphorylation:
Reduced electron carriers, produced by cytosolic dehydrogenases and mitochondrial oxidative pathways, become reoxidized by enzyme complexes bound in the inner membrane.
These complexes actively pump protons outward, creating an energy gradient whose discharge through complex V drives ATP synthesis.
二、Electron Transport
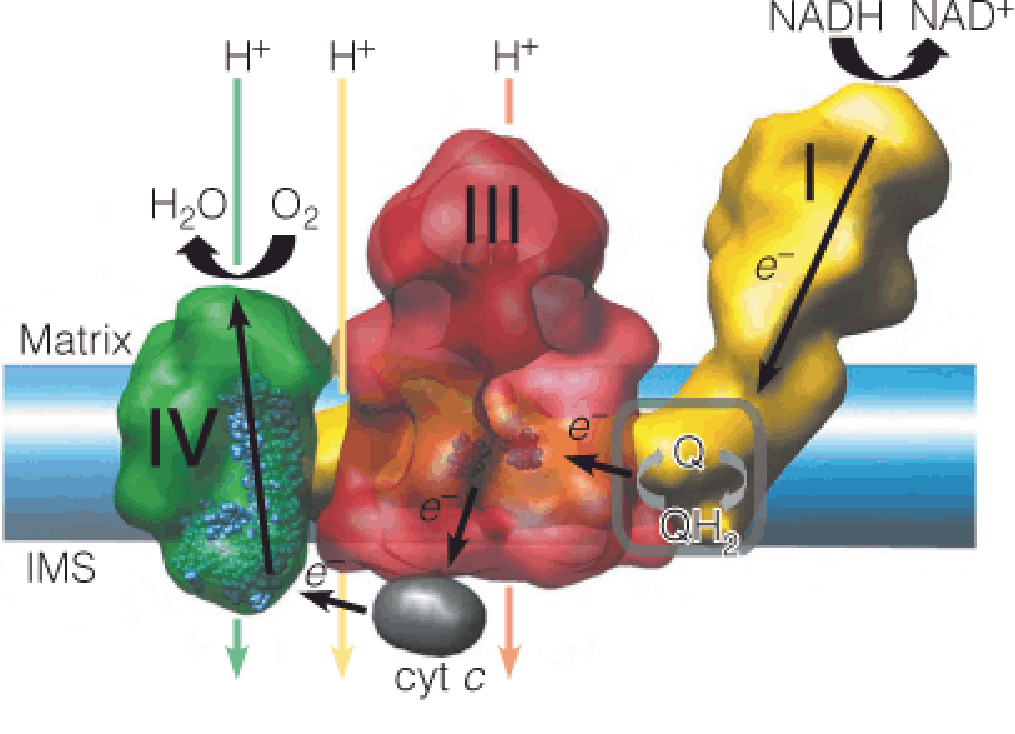
Embedded within the inner membrane are the protein carriers that constitute the respiratory chain.
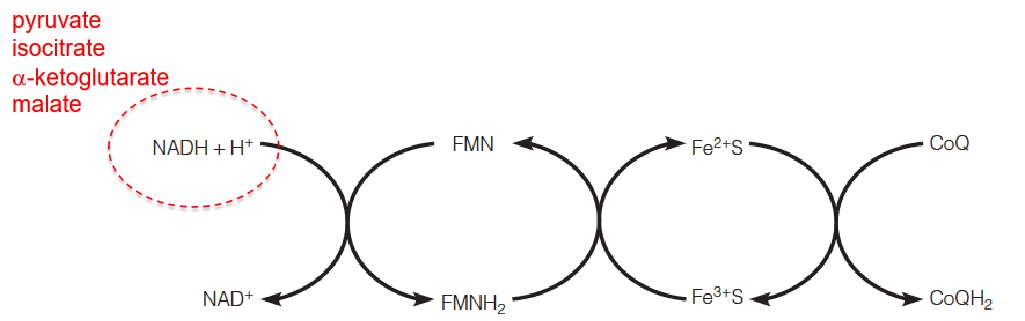
Complex I and complex II receive electrons from the oxidation of NADH and succinate, respectively, and pass them along to a lipid-soluble electron carrier, coenzyme Q, which moves freely in the membrane.
Complex III catalyzes the transfer of electrons from the reduced form of coenzyme Q to cytochrome c, a protein electron carrier that is also mobile within the intermembrane space (IMS).
Complex IV catalyzes the oxidation of cytochrome c, reducing O2 to water.
The energy released by these exergonic reactions creates a proton gradient across the inner membrane, with protons being pumped from the matrix into the intermembrane space.
- Protons then re-enter the matrix through a specific channel in complex V.
The energy released by this exergonic process drives the endergonic synthesis of ATP from ADP and inorganic phosphate.
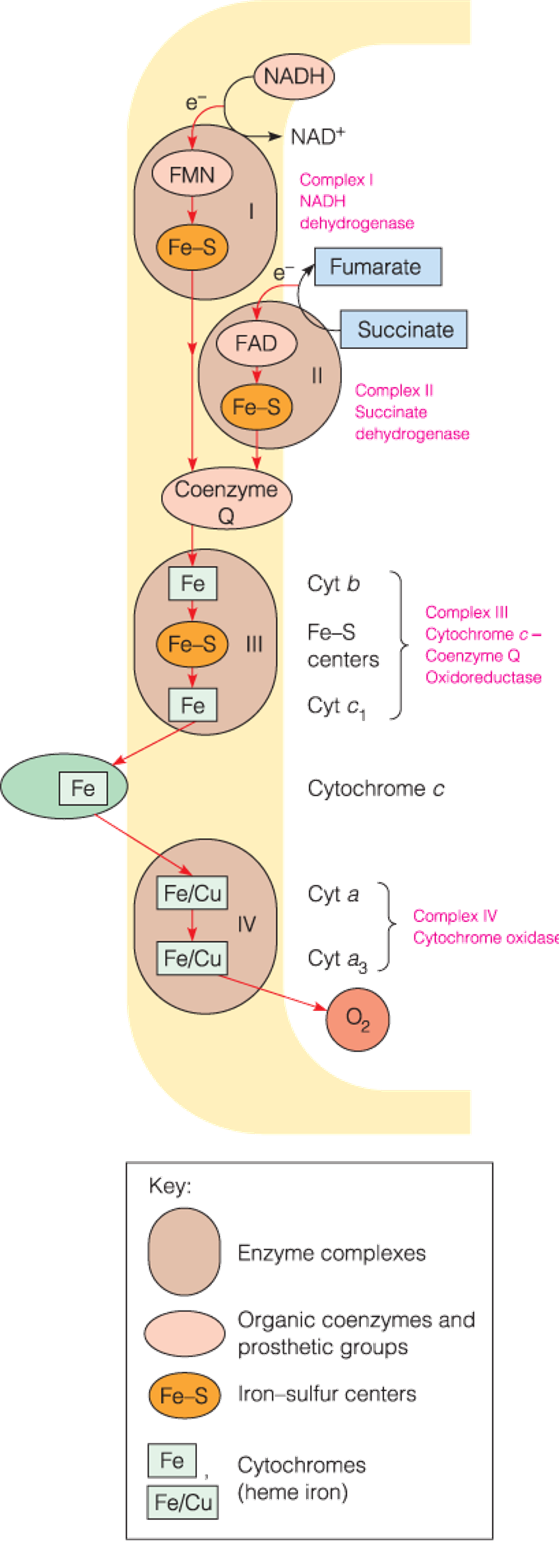
Respiratory electron carriers in the mitochondrion
- The sequence of electron carriers that oxidize succinate and NAD-linked substrates in the inner membrane.
- The respiratory chain catalyzes the transport of electrons from low-potential carriers to high-potential carriers.
Prokaryotic cells:
- Comparable(类似的) system
- Different electron carriers
- Inner membrane surface
- Electron transport and oxidative phosphorylation occur at the cell periphery(外围)
Electron Carriers in the Respiratory Chain
Flavoproteins (黄素蛋白)
- Contain tightly bound FMN(flavin mononucleotide,黄素单核苷酸) or FAD as redox cofactors.
- Each flavoprotein provides a different microenvironment for the isoalloxazine ring (异咯嗪环), conferring a unique standard reduction potential on the flavin.
- Flavin nucleotides can act as transformers between two-electron and one-electron processes due to their ability to exist as stable one-electron reduced semiquinone(半醌) intermediates.
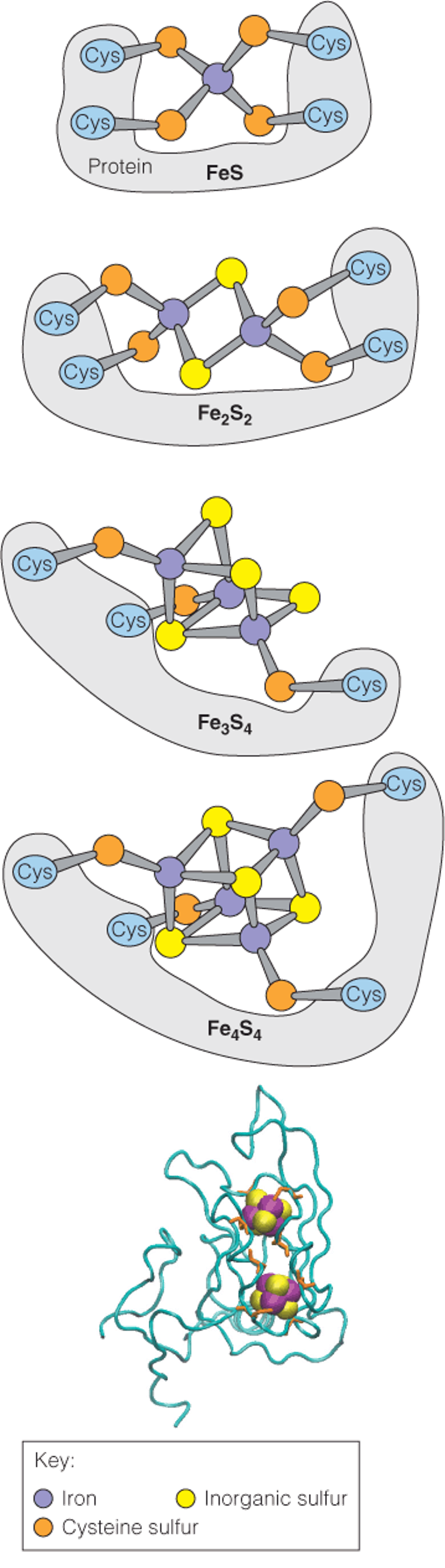
Iron–sulfur proteins
- Iron–sulfur clusters consist of nonheme iron complexed to sulfur in four known ways
Coenzyme Q
Cytochromes
1. Structures of iron–sulfur clusters
The top panel illustrates the covalent attachment of a pair of clusters in a subunit of complex I from Thermus thermophilus (嗜热菌)
2. Coenzyme Q
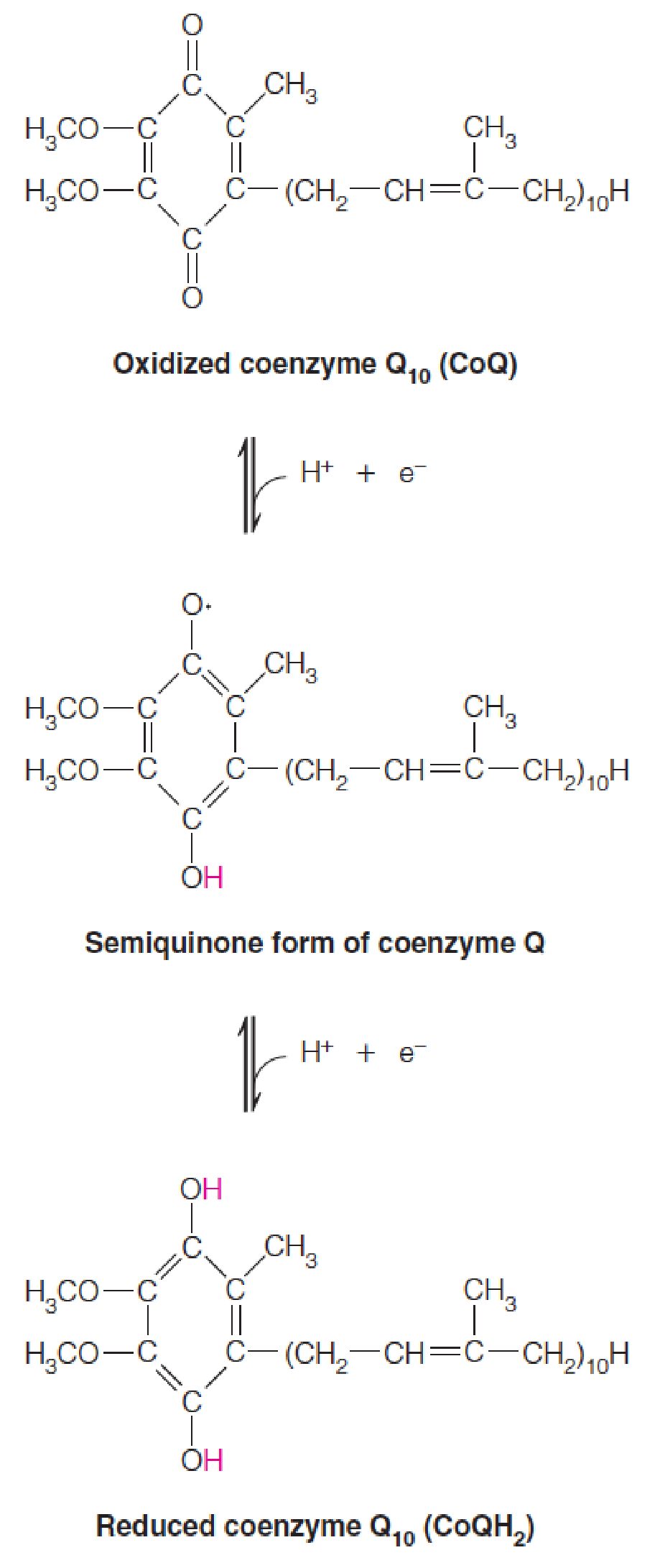
The cytochrome Q carrier was found to be a benzoquinone (苯醌) linked to a number of isoprene units, usually 10 in mammalian cells and 6 in bacteria.
Because the substance is ubiquitous (泛醌) in living cells, one group of researchers named it ubiquinone, while another called it coenzyme Q, or Q.
- Extremely lipophilic (亲脂性的)
- Diffuse rapidly through the inner mitochondrial membrane;
- One electron at a time (like FAD and FMN)
- Two-electron carriers
3. Cytochromes
Absorption spectra of cytochromes
The plots show the absorption spectra of cytochromes b, c, and a in their oxidized (red) and reduced (blue) states.
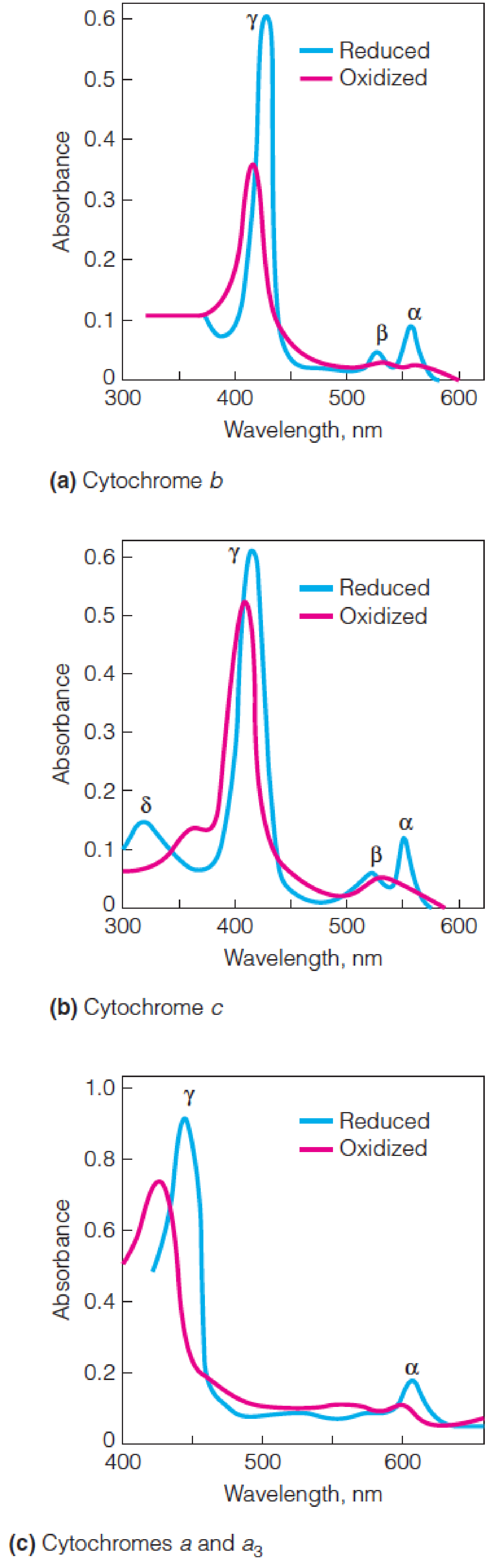
- Cytochrome b from Neurospora (脉孢霉)
- Cytochrome c from horse heart
- Cytochrome oxidase from bovine(牛的) heart
- Contains both cytochromes a and a
3.
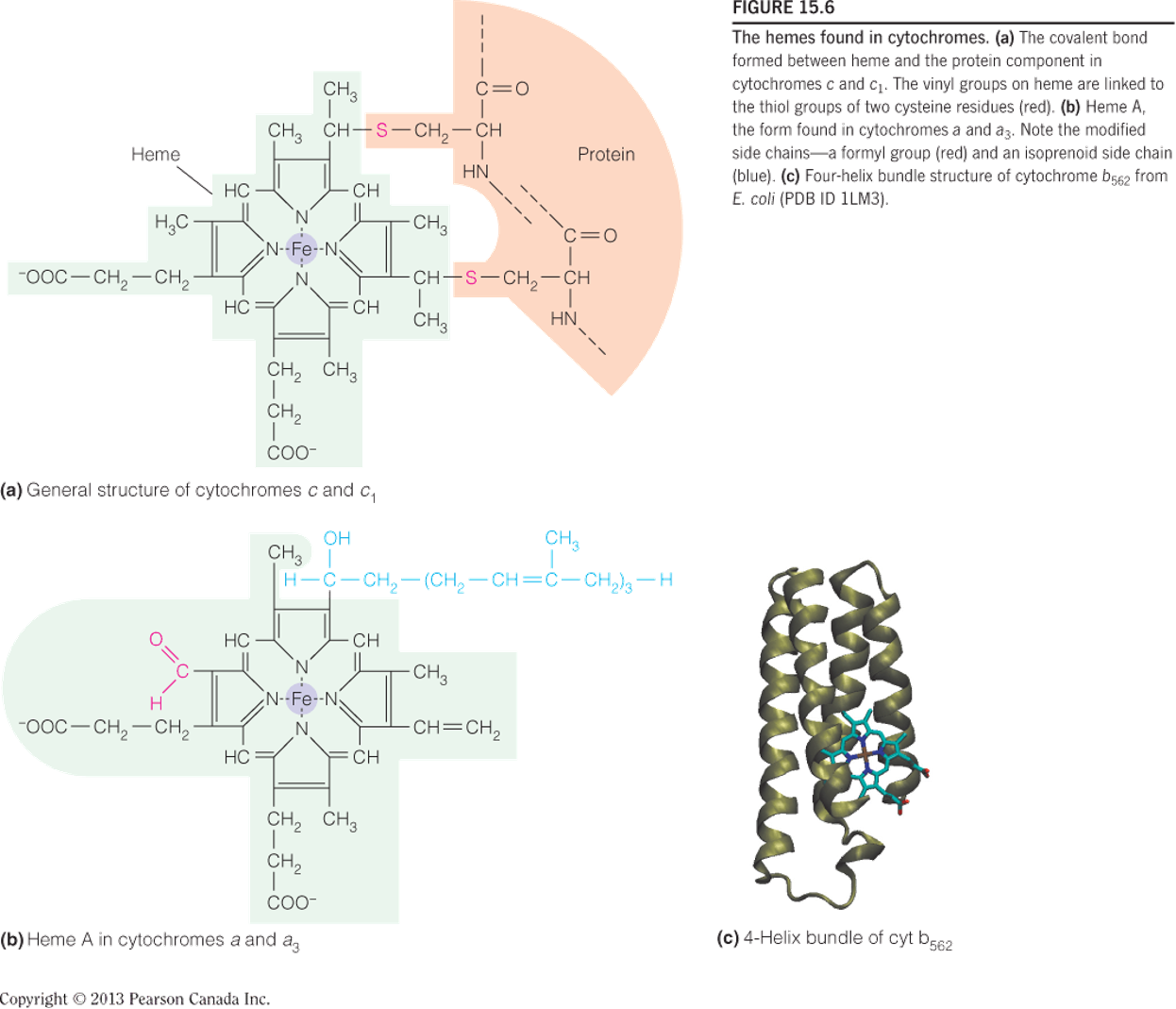
The hemes found in cytochromes
cytochromes 中的 heme:非血红素铁
The covalent bond formed between heme and the protein component in cytochromes c and c
1.- The vinyl groups (乙烯基) on heme are linked to the thiol groups of two cysteine residues (red).
Heme A, the form found in cytochromes a and a3. Note the modified side chains—a formyl group (red) and an isoprenoid side chain (blue).
Four-helix bundle structure of cytochrome b562 from E. coli
Determining the sequence of respiratory electron carriers
Standard reduction potentials (标准还原电位) of the major respiratory electron carriers:
Three reactions in the respiratory chain have $\Delta G ^{\circ}$ values greater than -30.5 kJ/mol (课本P629), the $\Delta G ^{\circ}$ for ATP hydrolysis:
$$
FMN \rarr Q; \ cyt\ b \rarr cyt\ c_{1}; and \ cyt\ a \rarr O_{2}
$$
Reduction potential 的排列:
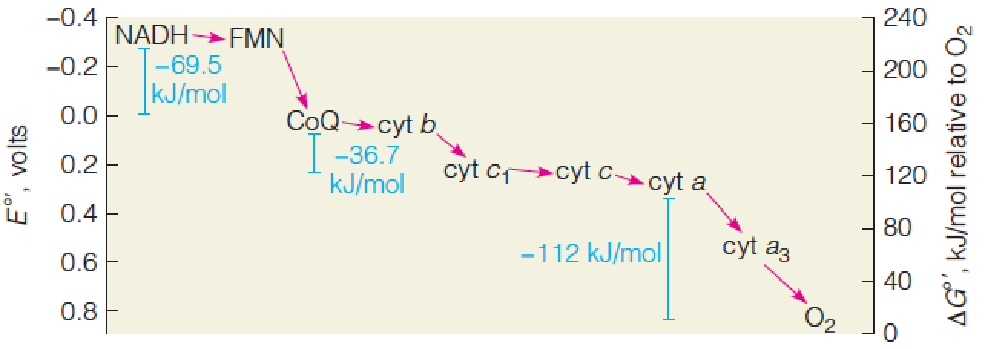
Electrons are transferred from low-potential to high-potential carriers
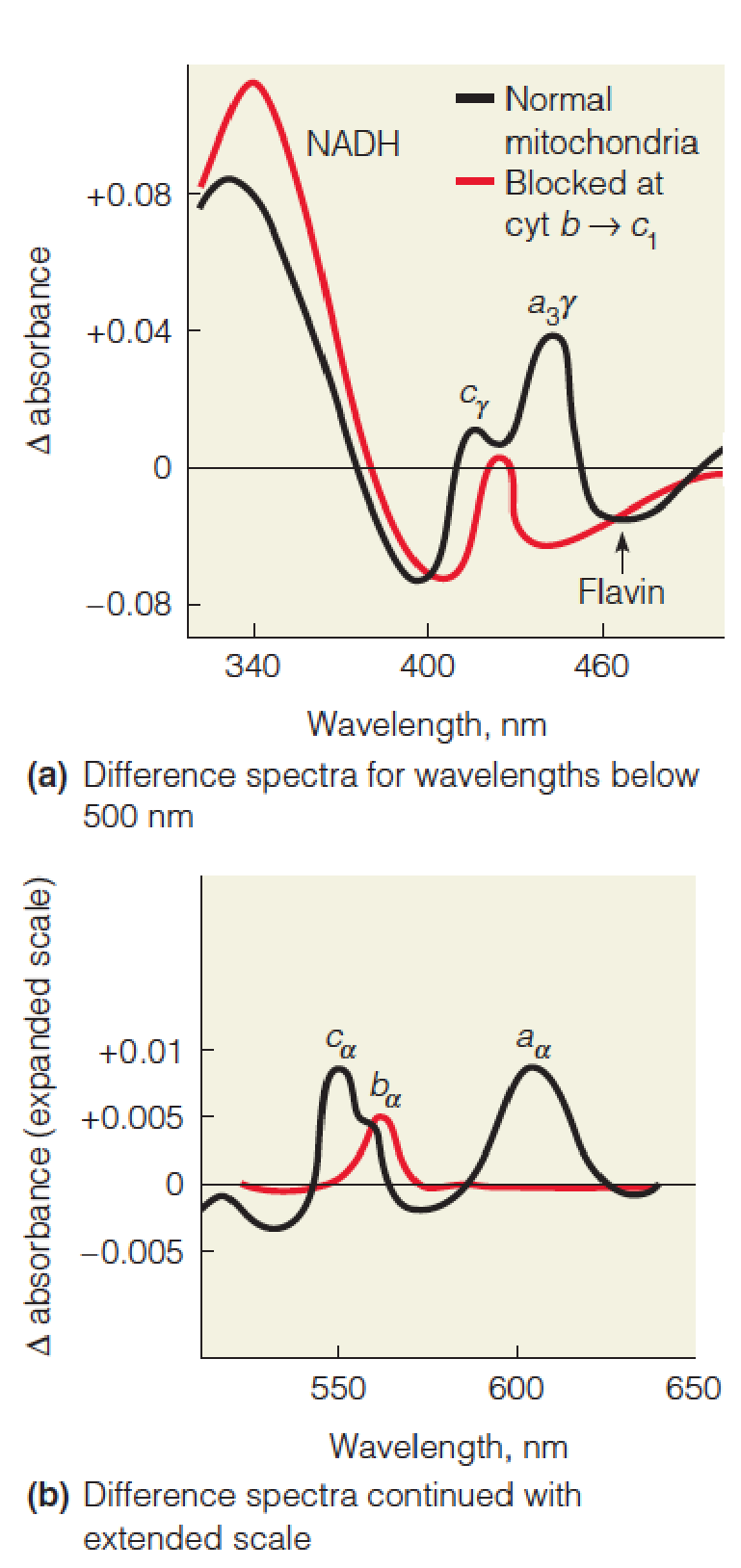
Difference spectra (差分光谱) of mitochondria
These difference spectra of rat liver mitochondria were recorded in a double-beam spectrophotometer (双光束分光光度计).
The black line shows the difference spectrum of fully reduced versus fully oxidized mitochondria.
Mitochondria reduced with substrate under anaerobic conditions were in the sample chamber, and oxygenated mitochondria in the reference chamber.
The peaks and shoulders refer to NADH, flavin, and the α and γ absorption bands, as indicated, for cytochromes a, a3, b, and c.
The red line shows the effect of adding a respiratory inhibitor, antimycin A, which blocks electron flow from cytochrome b to c1.
The inhibitor causes all of the carriers beyond cytochrome b to become fully oxidized, while NADH, flavin, and cytochrome b are in the reduced state. Note the expanded absorbance scale beyond 500 nm.
Sites of action of some respiratory inhibitors and artificial electron acceptors
This schematic of the respiratory chain from NADH to O2 shows the sites of action of some useful inhibitors (red) and some artificial electron acceptors (blue).
Each acceptor is positioned according to its value (in parentheses), identifying the most likely site at which an acceptor will withdraw electrons from the respiratory chain when added to mitochondria.
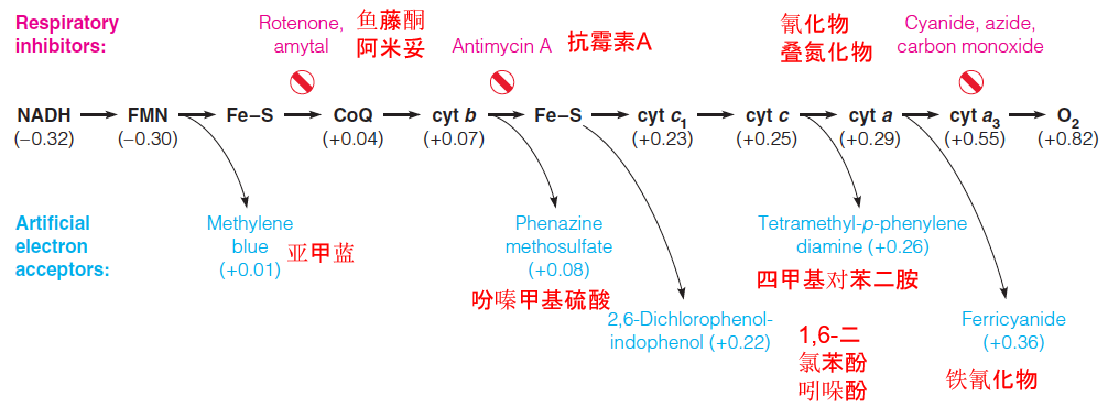
All of the carriers upstream of the inhibition site become reduced;
All of the carriers downstream of the inhibition site become oxidized
The order of action of electron carriers in the respiratory chain was elucidated by:
- difference spectrophotometry 差分光度法
- analysis of respiratory inhibitors
- properties of membrane complexes.
Mitochondrion can be disrupted by mechanical treatment, such as sonication; by detergent digitonin(洋地黄皂苷) to remove outer membrane; 5 complexes
Electron transfer activity is retained
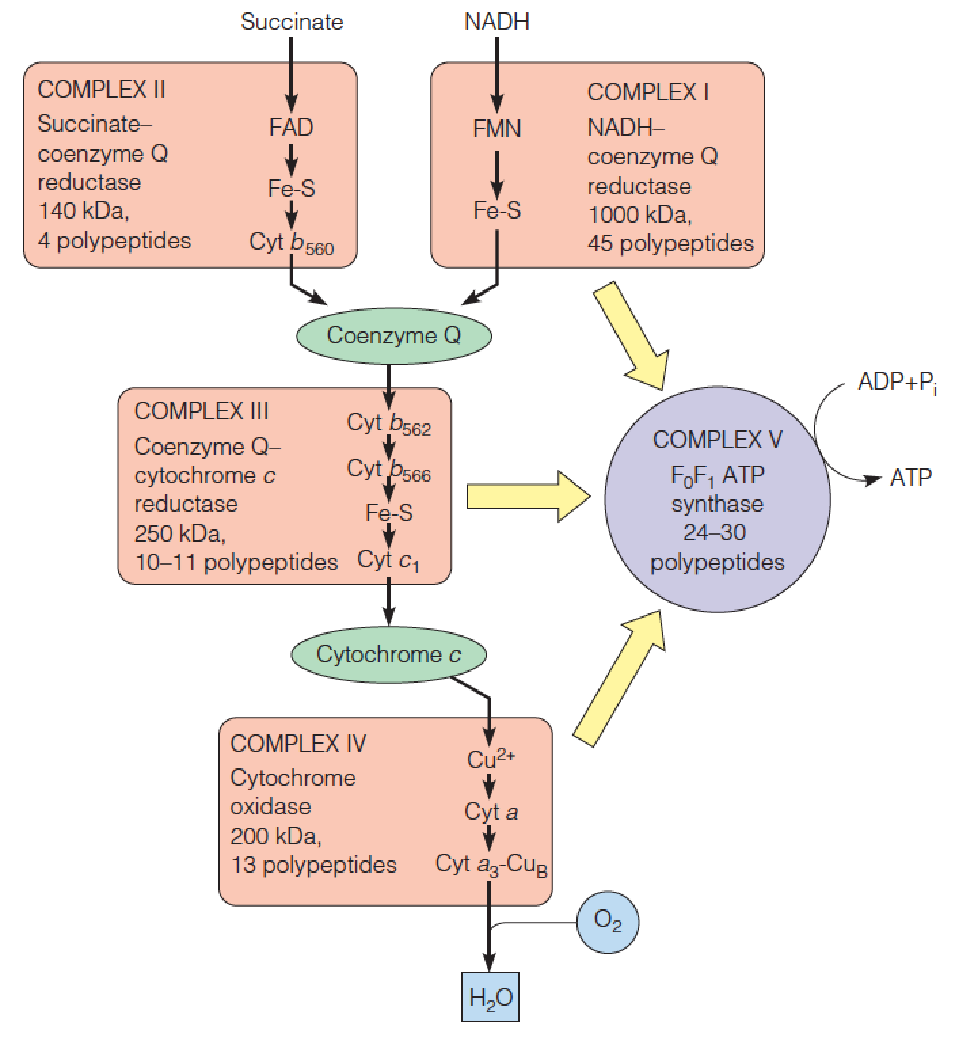
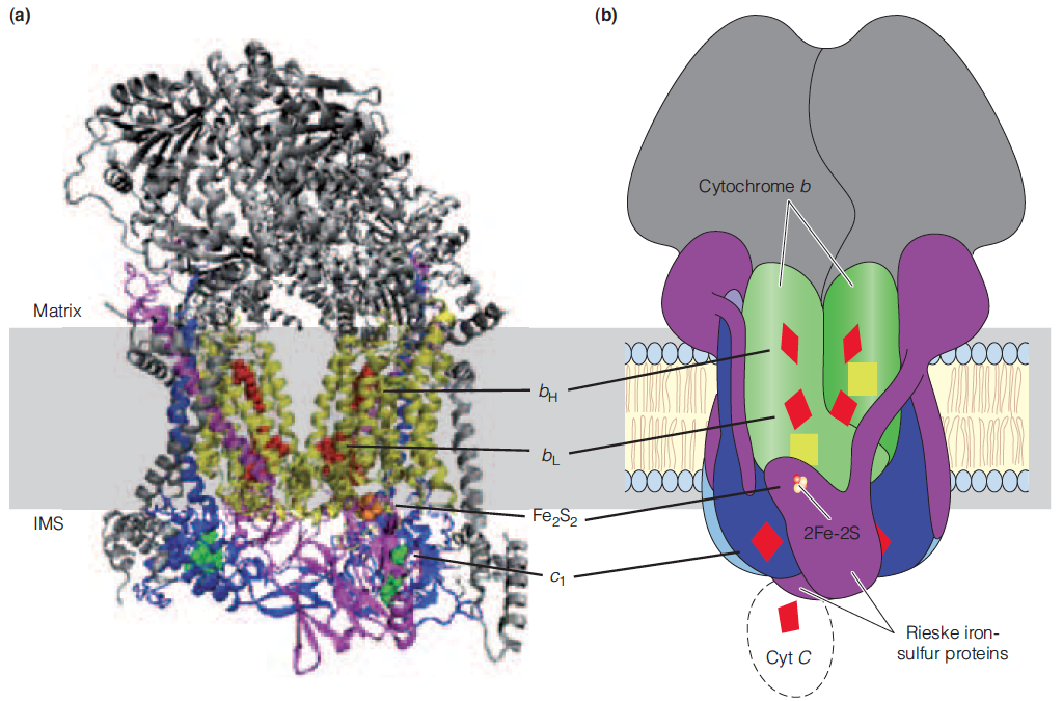
Multiprotein complexes in the mitochondrial respiratory assembly
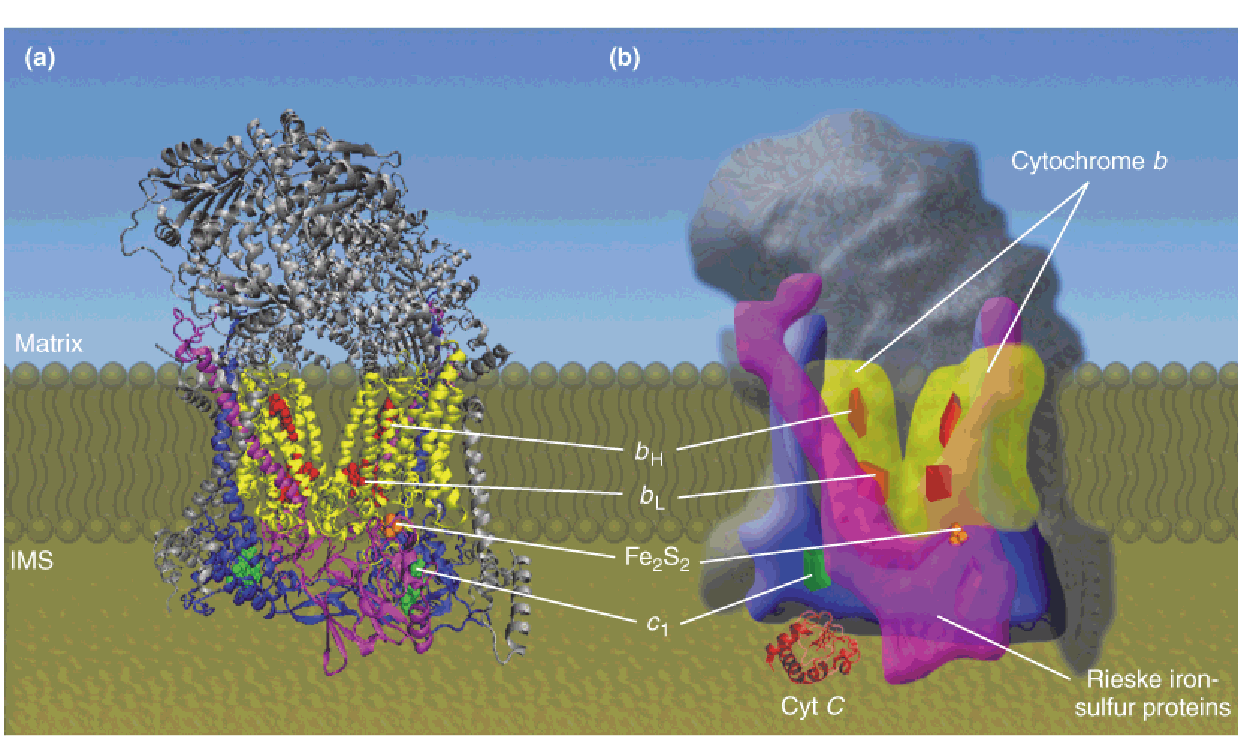
The subscripts for the b cytochromes denote their spectral maxima.
The two b hemes in complex III, identified as cyt b562 and cyt b566, are bound to the same polypeptide chain.
The yellow arrows denote the energy released by the actions of complexes I, II, and IV used to drive the synthesis of ATP by complex V (ATP synthase).
1. Complex Ⅰ
The overall reaction catalyzed by the NADH dehydrogenase complex (complex I ) is shown below.
Because the electrons are subsequently used to reduce coenzyme Q, a more descriptive name for this complex is NADH–coenzyme Q reductase.
Electron transport through complex I is coupled to the pumping of protons from the matrix to the intermembrane space.
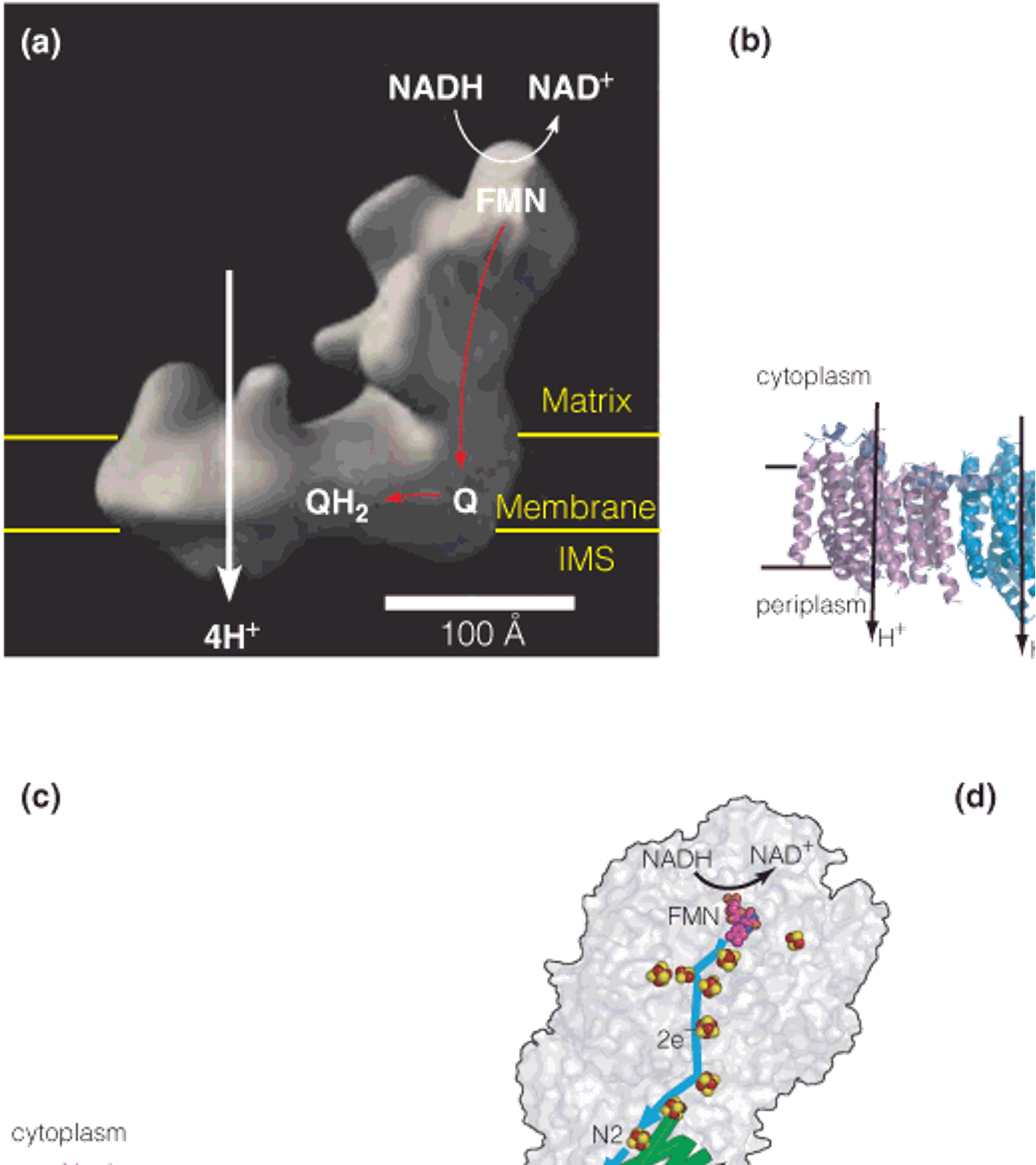
Structure and function of complex I (NADH–coenzyme Q reductase)
Model of complex I from the yeast Yarrowia lipolytica (解脂耶氏酵母)
X-ray structure of the entire complex I from the archaea Thermus thermophilus (嗜热菌).
Proposed model of proton translocation by complex I. NADH binds near FMN.
Proposed model for coupling of electron transport to proton pumping by complex I.
- NADH dehydrogenase
NADH is the main donor of electrons into the respiratory chain
Numerous dehydrogenases
NADH is readily disassociated from enzymes, soluble cofactor (Redox), carrying electrons
NADH + H^+^ + Q ⟶ NAD^+^ + QH2
Complex I pumps protons.
2. Complex Ⅱ
Structure of complex II (succinate dehydrogenase)
The enzyme is composed of two hydrophilic subunits extending into the matrix, the FAD-binding subunit (blue) and the iron–sulfur subunit (yellow), and two transmembrane subunits (pink and gold).
The path of electron transport from FAD through the three iron–sulfur clusters to coenzyme Q (UQ) is shown on the right.
Succinate binds near FAD in the blue subunit.
On inner mitochondrial membrane (IMM), not in the matrix like all other citric acid cycle enzymes;
Complex II does not pump protons
Succinate + Q ⟶ fumarate + QH2
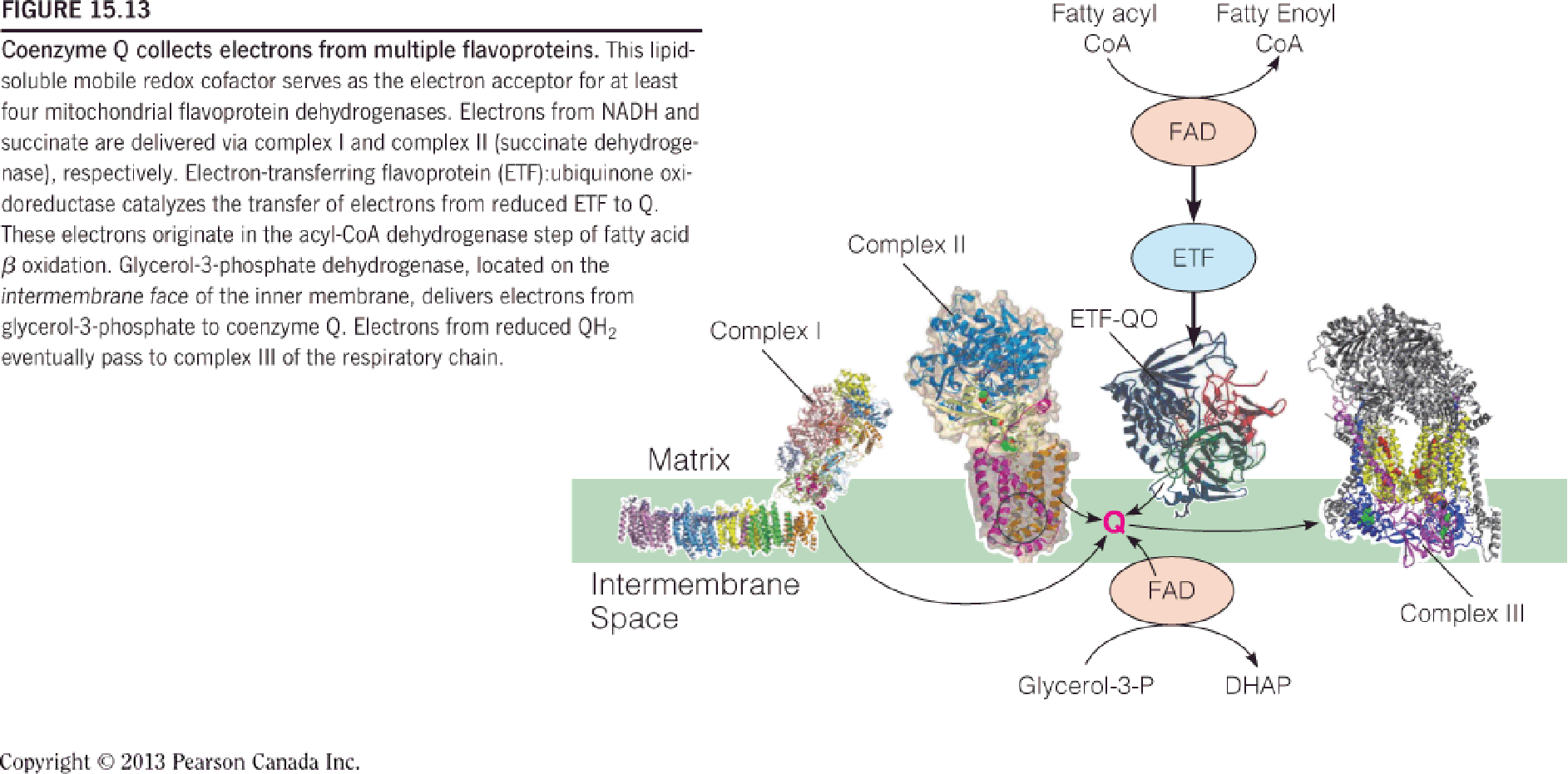
CoQ is lipid-soluble, moving fast in the lipid bilayer membrane;
CoQ draws electrons from (CoQ可以接收多个渠道的电子):
- NADH dehydrogenase (complex I);
- succinate dehydrogenase (complex II);
- fatty acid dehydrogenase;
- G3P dehydrogenase;
CoQ is subsequently oxidized by complex III;
G3P→DHAP; shuttling cytoplasmic NADH to mitochondrion;
3. Complex Ⅲ
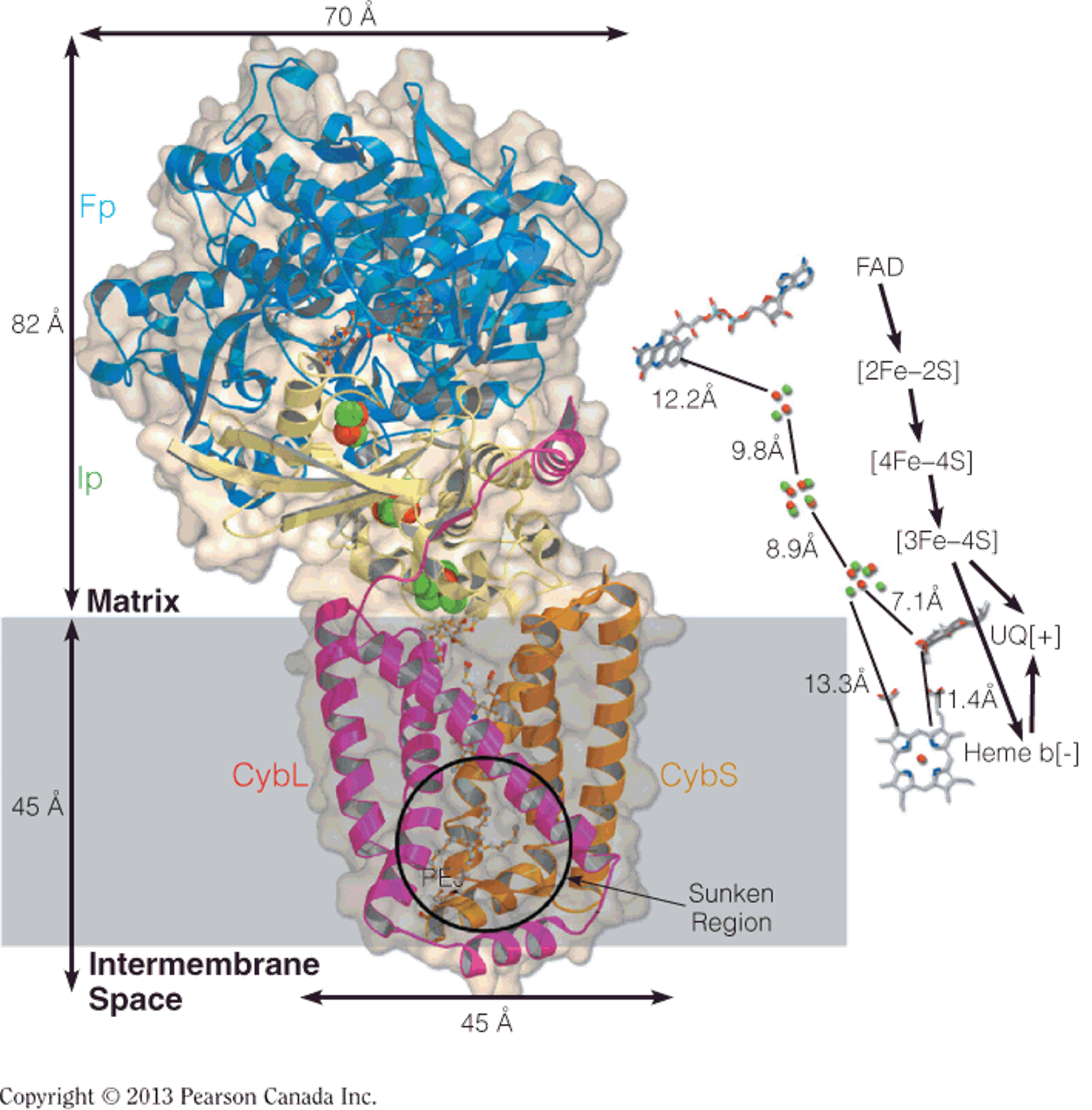
Structure of Complex III (coenzyme Q:cytochrome c oxidoreductase(氧化还原酶))
X-ray structure of the dimeric complex
Cartoon of the complex III dimer illustrating the arrangement of the subunits and redox carriers.
- The approximate location of the complex in the inner membrane is indicated.
Cyt c, a peripheral protein electron carrier, mobile within the intermembrane space (IMS).
Transfer electrons from QH2 to cyt c (between complexes III and IV)
Dimer each monomer: 10-11 proteins
Cyt c: apoptosis
Q cycle: explain the stoichiometry
- QH2: two electron donor
- Cyt: one electron donor
4. Complex Ⅳ
Structure of cytochrome c oxidase (complex IV)
Four electrons are donated, one at a time, by reduced cyt c, to the CuA center, through heme a, and on to the catalytic site (binuclear a3–CuB site) where one molecule of O2 is reduced, yielding two molecules of H2O.
Protons are pumped from the matrix side to the intermembrane space (IMS) side of the membrane.
The final stage of electron transport
Large homodimeric complex
Transfer electrons from reduced cyt c, through a, a3, to O2
For electron transferred, complex IV pumps one proton from matrix to IMS
Model of proposed supercomplex of complexes I, III, and IV:
三、Oxidative Phosphorylation
Experimental identification of “coupling sites.”
Electron transport is restricted to particular parts of the chain by use of selected electron donors, electron acceptors, and respiratory inhibitors, as indicated.
For each segment of the chain that is thus isolated, P/O ratios are determined, allowing identification of coupling sites.
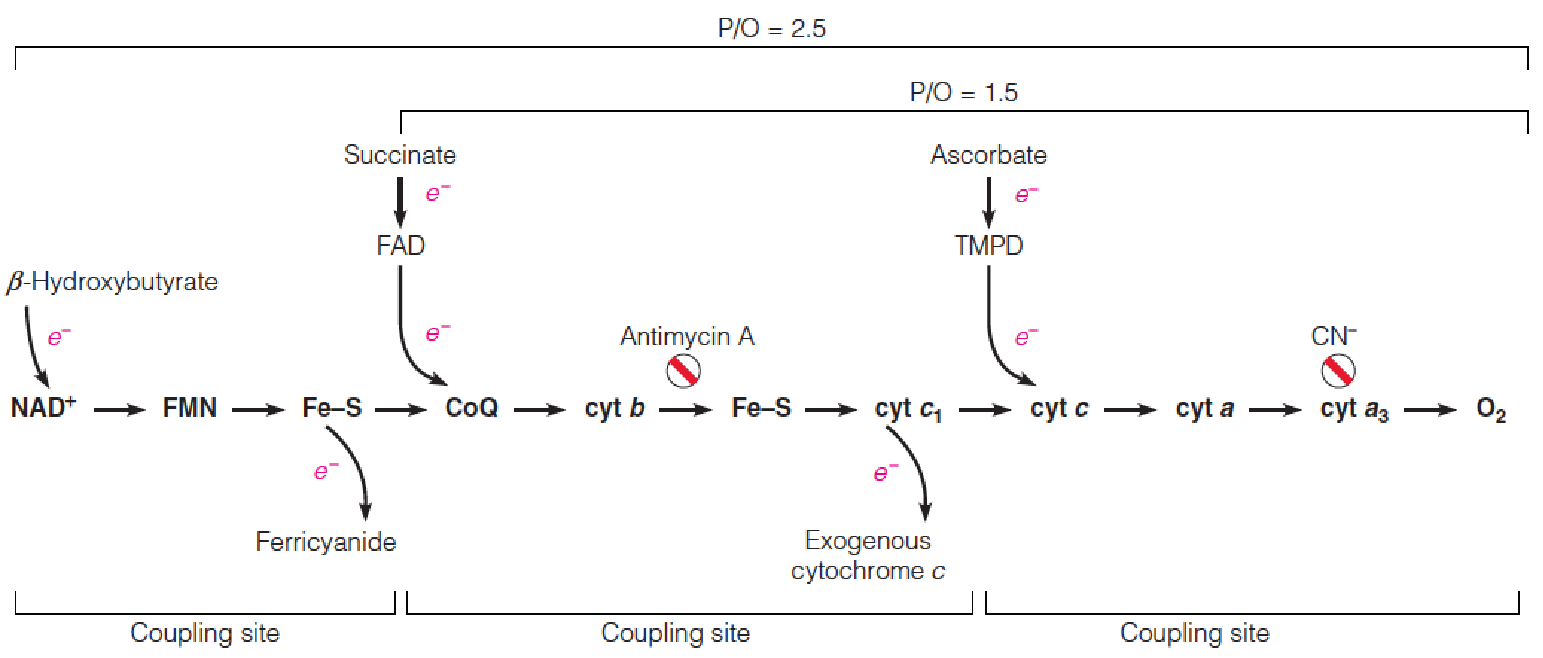
ATP synthesis is quantified by phosphorylation (Pi used); electron pairs are measured as oxygen uptake
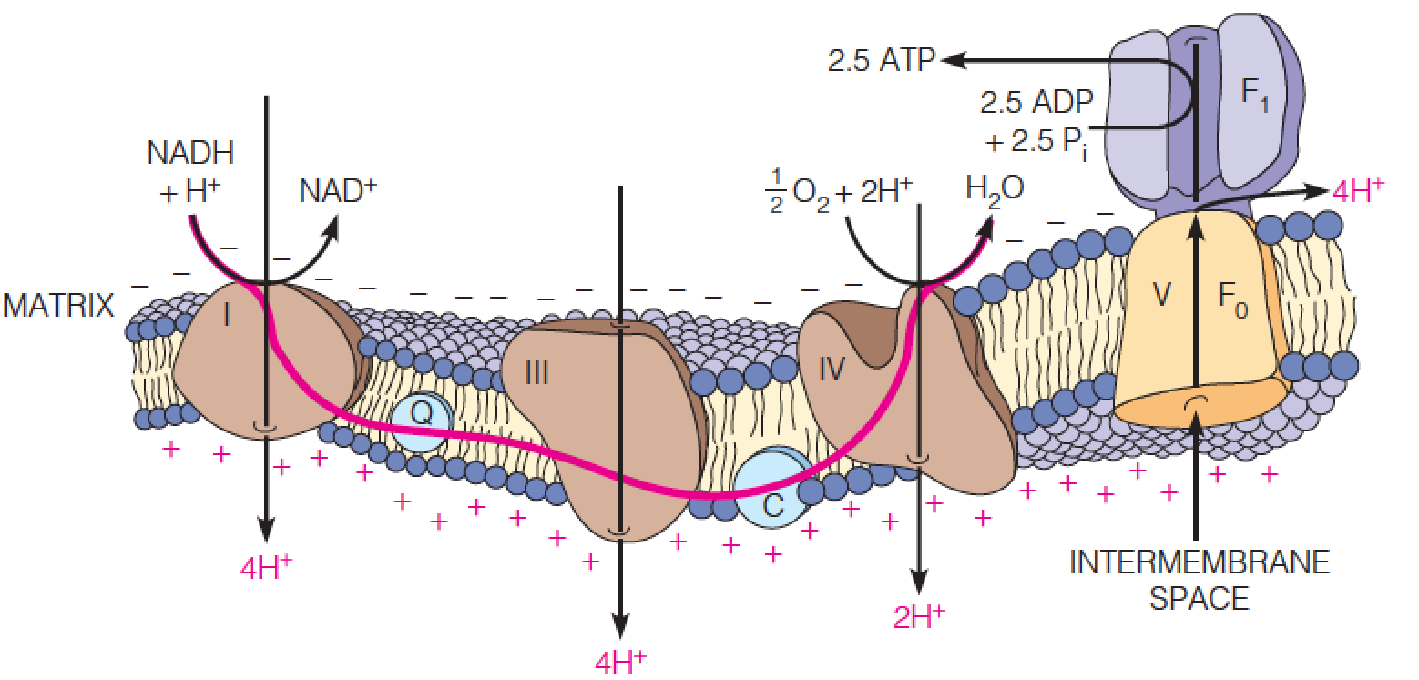
Chemiosmotic (化学渗透的) coupling of electron transport and ATP synthesis
Protons are pumped by complexes I, III, and IV as electrons flow through the complexes, generating an electrochemical gradient across the membrane (protonmotive force, pmf, 质子驱动力).
Proton re-entry to the matrix, through the F0 channel of ATP synthase (complex V), provides the energy to drive ATP synthesis.
Chemiosmotic coupling(化学渗透耦联) refers to the use of a transmembrane proton gradient to drive endergonic processes like ATP synthesis.
“direct chemical coupling”: no
Peter Mitchell (1961), British biochemist Nobel Prize in 1978
Oxidation and phosphorylation are closely coupled!
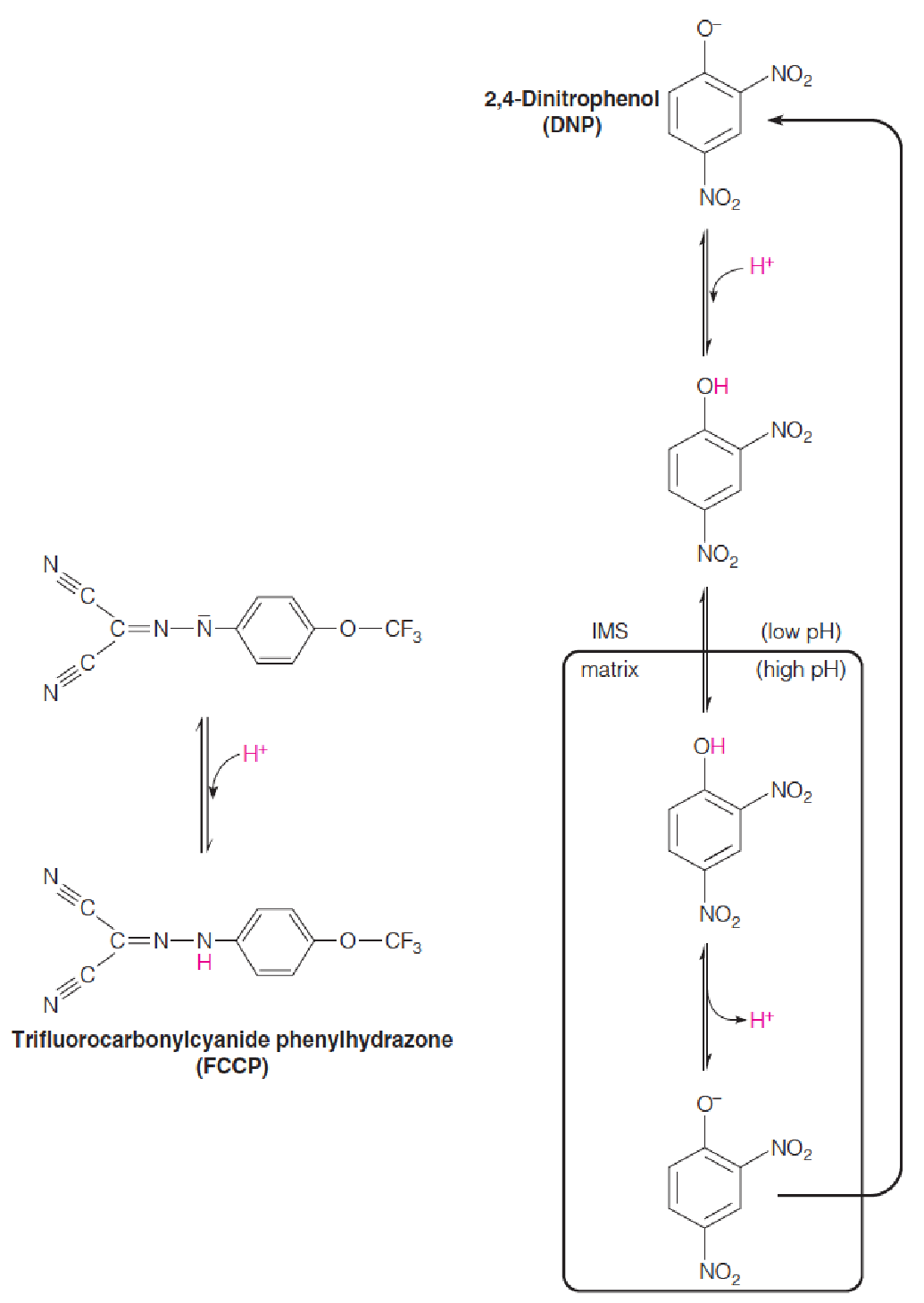
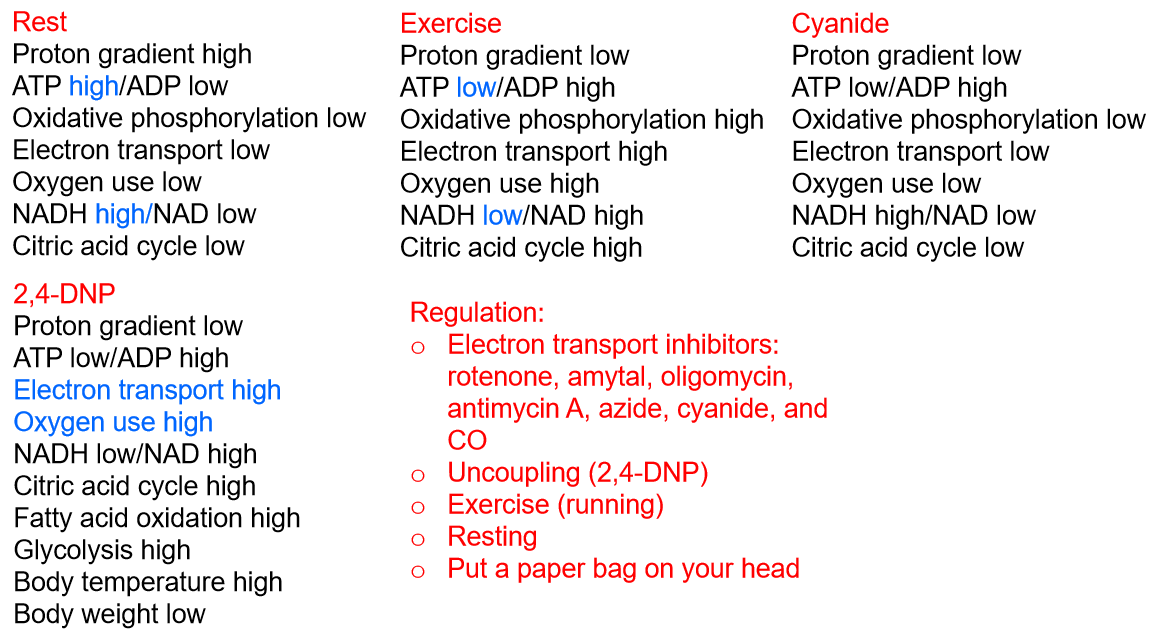
1. Uncouplers Act by Dissipating the Proton Gradient
The lipophilic weak acids 2,4-dinitrophenol (DNP, 2,4-二硝基苯酚) and trifluorocarbonylcyanide phenylhydrazone (FCCP, 三氟羰基氰苯腙) are called uncoupling agents, or uncouplers (解耦器).
Uncoupling agents, when added to mitochondria, permit electron transport along the respiratory chain to O2 to occur in the absence of ATP synthesis.
They uncouple the process of electron transport from the process of ATP synthesis. 破坏H+ gradient,电子传递仍在进行,但是ATP不进行合成。
dieting drug: kill people
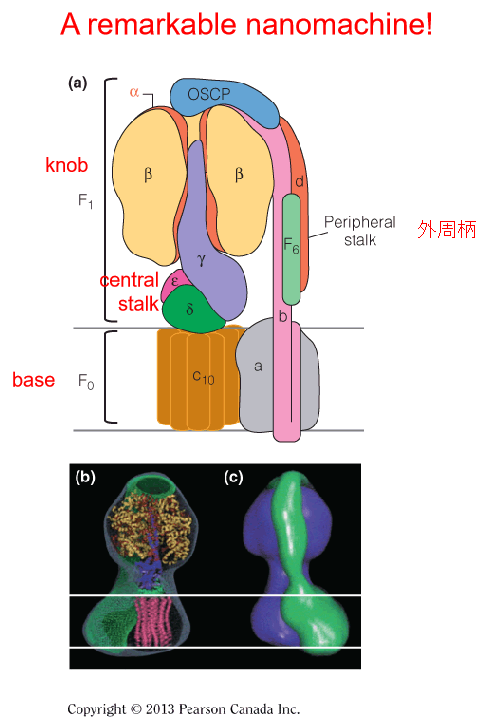
2. Structure of the F0F1 complex (Complex Ⅴ)
The F0F1 complex, also called ATP synthase or complex V, contains an F1 knob projecting into the mitochondrial matrix and connected by a central stalk to the F0 base.
The globular F
1knob contains three $\alpha \beta$ dimers, arranged above the central stalk, which is made up of γ, δ and ε subunits.The F
0base is composed of 10–12 c subunits (the c-ring) and one subunit a. The peripheral stalk (subunits b, d, F6, and OSCP) is attached to the F0base via subunit a and at least four other minor membrane-embedded subunits that are not shown.
The central stalk and the c-ring compose the “rotor(转子)” of ATP synthase.
The remainder of the subunits make up the “stator(定子),” a structure that prevents the rotation of the three dimers of F1.
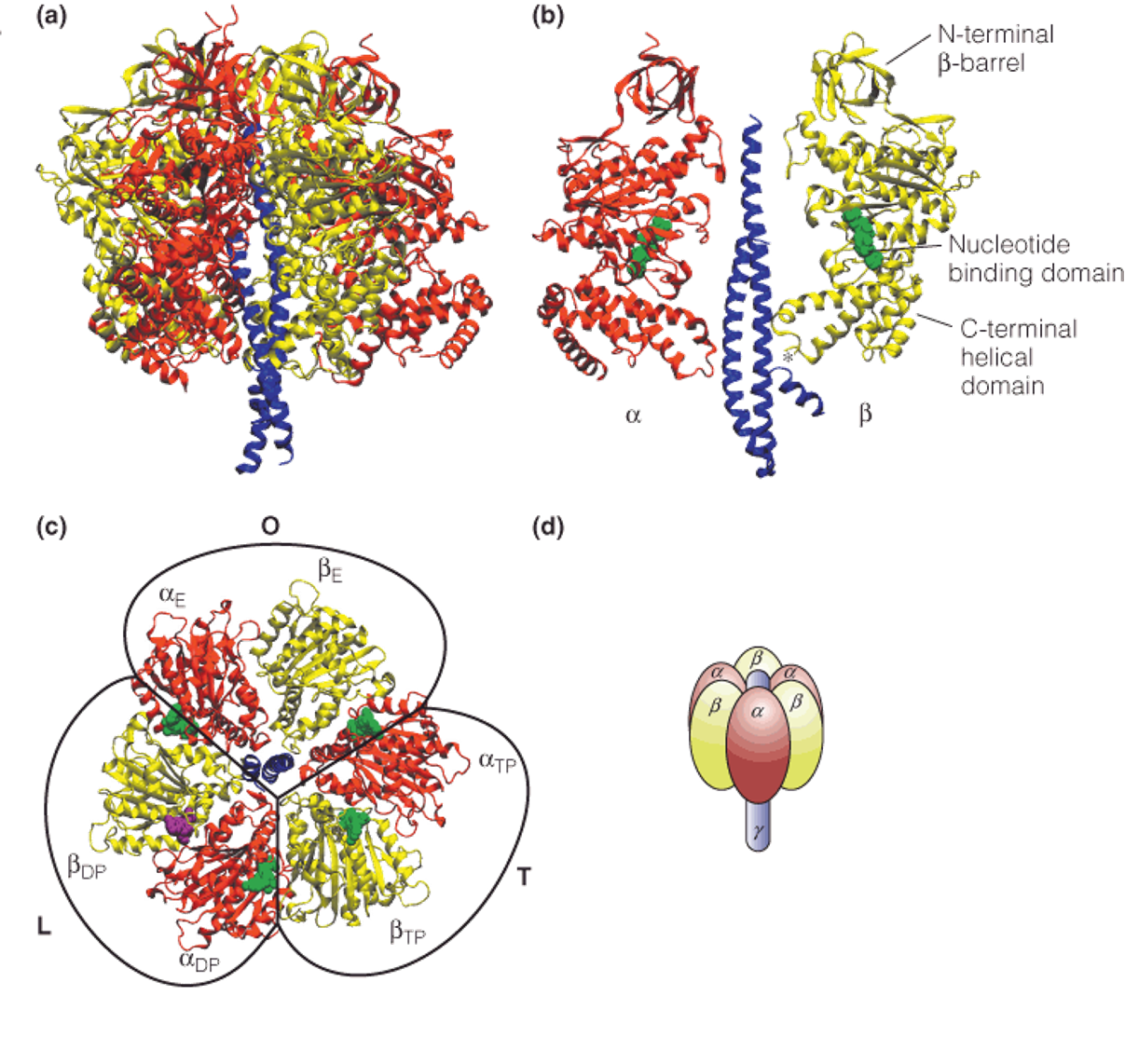
3. Structure of the mitochondrial ATP synthase complex
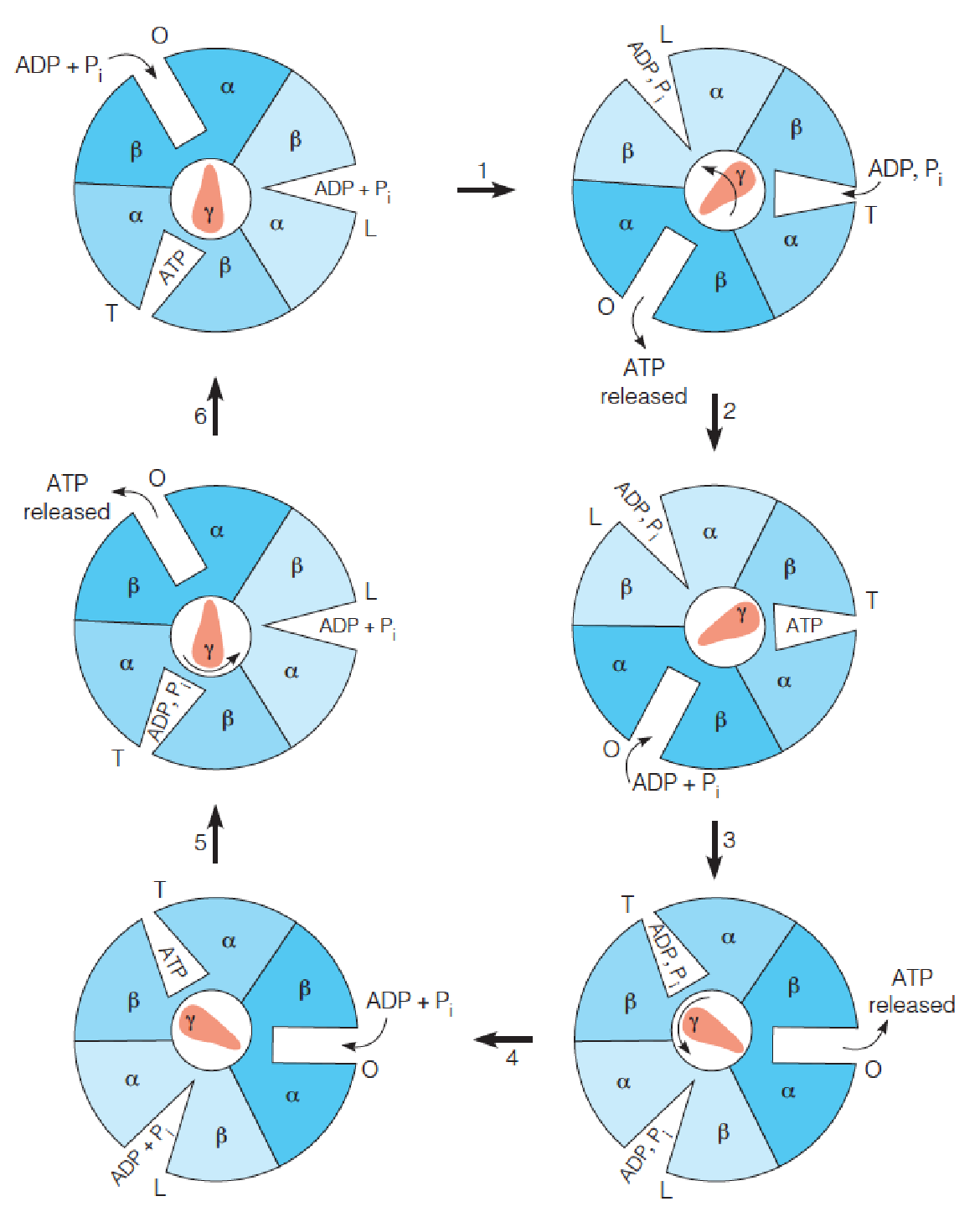
Binding change model for ATP synthase
The nucleotide binding site (catalytic site) of the three dimers exists in three different conformations, termed loose (L), tight (T), and open (O).
The subunit rotates counterclockwise, driven by the passage of protons through channels in F0, while the dimer assemblies are held stationary by a stator.
Step 1 represents 120° rotation of the subunit, leading to a conformational change in all three dimers, such that the T site changes to an O site, leading to ATP release, and an O site changes to an L site, binding ADP and Pi.
The third site changes from an L conformation, with loosely bound ADP and Pi, to a T conformation, where the substrates are tightly bound, leading to ATP formation in step 2.
The experimental system that permits observation of rotation in the F1 component of F1F0 ATP synthase
The cloned gene encoding the F1b subunit was modified by adding a sequence that encodes an oligohistidine sequence that binds to a nickel-coated bead (Ni-NTA coated bead) in the conformation shown.
Thus, the complex was immobilized on the bead and attached to a glass coverslip.
Streptavidin is a protein used to couple fluorescent-tagged actin to the subunit.
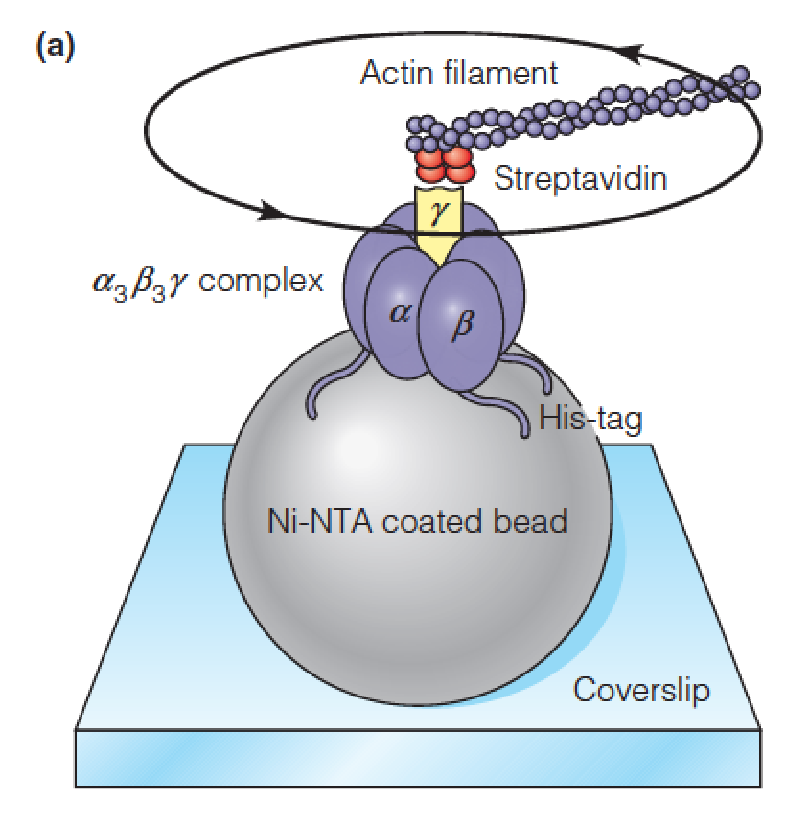
Fluorescence microscopic examination showed that, following addition of ATP and its subsequent hydrolysis by the catalytic subunits, the actin molecule was rotating, which proved that the subunit itself was rotating.
The time interval between images is 133 ms.
4. Proton-driven rotation of the c-ring of the component of ATP synthase
This model is based on the X-ray structure of the E. coli ATP synthase.
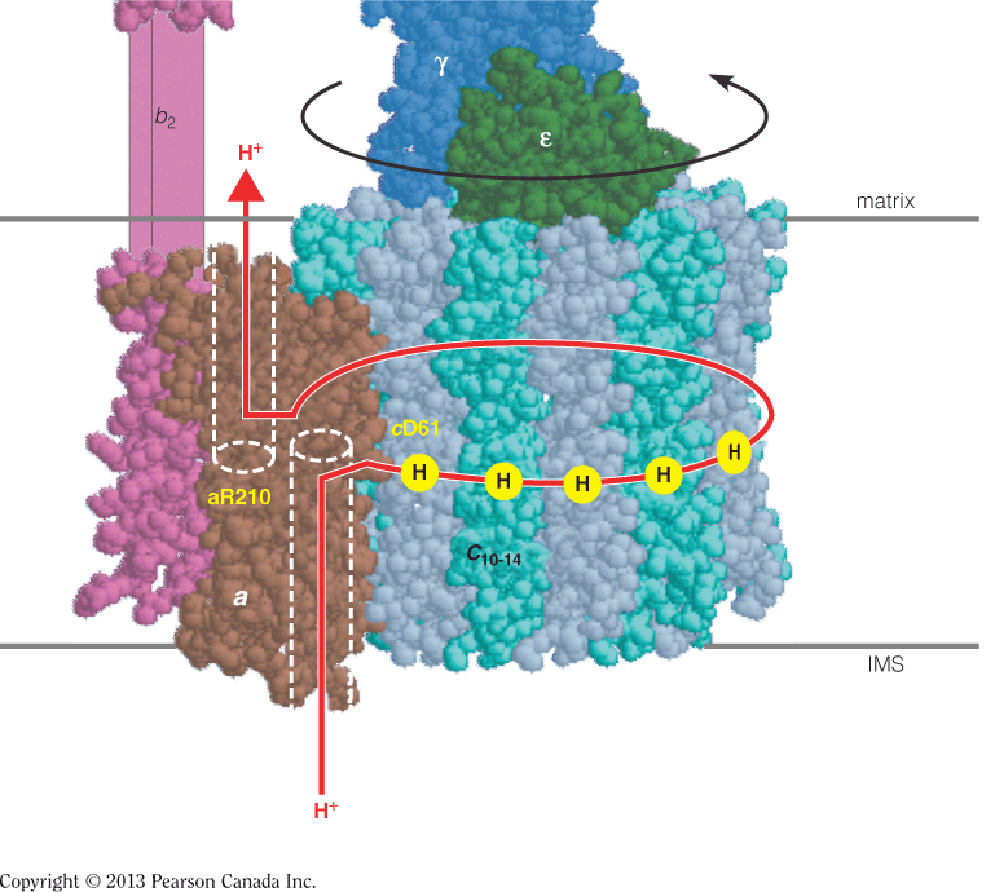
No ADP: stops it
No proton gradient: stops it
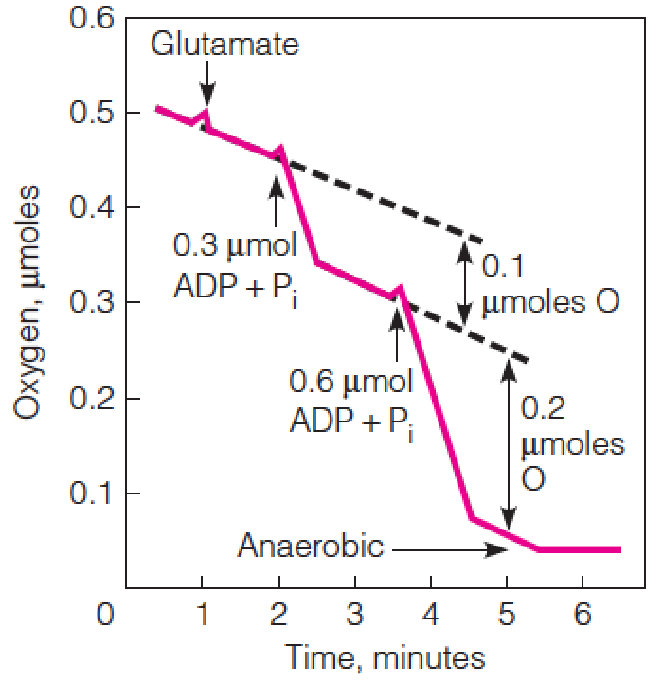
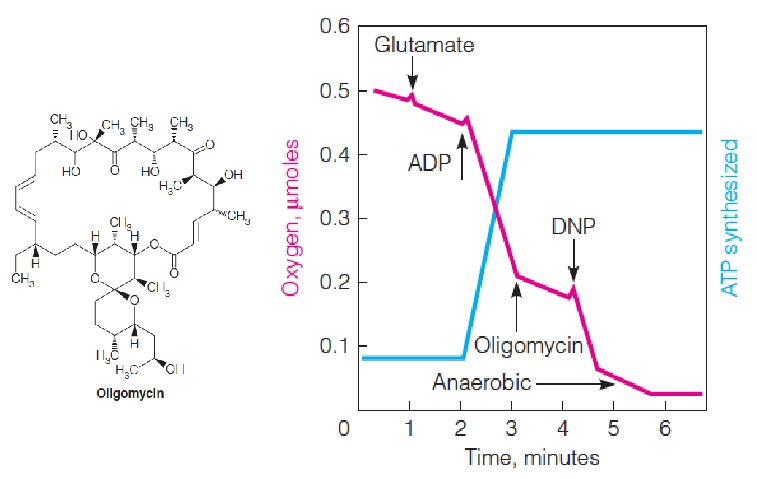
Experimental demonstration of respiratory control
Oxygen uptake is monitored in carefully prepared coupled mitochondria.
The addition of an exogenous oxidizable(可氧化的) substrate (glutamate) stimulates respiration only slightly, unless ADP + Pi is added as well.
Both ADP additions represent limiting amounts; the second addition is twice the amount of the first, to show that the magnitude of oxygen uptake is stoichiometric.
The slow oxygen uptake at the beginning results from endogenous substrates in the mitochondria.
ADP stimulates respiration only until all of the ADP + Pi has been converted to ATP.
Oxygen uptake is recorded in moles O, because one pair of electrons reduces one atom of O, not one molecule of O2.
加入ADP后,O2的消耗显著增多,之后又恢复正常,后加入2倍浓度的ADP,消耗量大致也提升2倍:
In most aerobic cells the level of ATP exceeds that of ADP by 4- to 10-fold.
It is convenient to think of respiratory control as a dependence of respiration on ADP as a substrate for phosphorylation.
If the energy demands on a cell cause ATP to be consumed at high rates, the resultant accumulation of ADP will stimulate respiration, with concomitant activation of ATP resynthesis.
Conversely, in a relaxed and well-nourished cell, ATP accumulates at the expense(开支,消费) of ADP, and the depletion of ADP limits the rate of both electron transport and its own phosphorylation to ATP.
Thus, the energy-generating capacity of the cell is closely attuned (协调) to its energy demands.
Effects of an inhibitor and an uncoupler on oxygen uptake and ATP synthesis.
DNP uncouples respiration from phosphorylation so that O2 uptake is stimulated even in the presence of oligomycin (ATP synthase inhibitor), but ATP synthesis remains blocked.
Respiratory Control
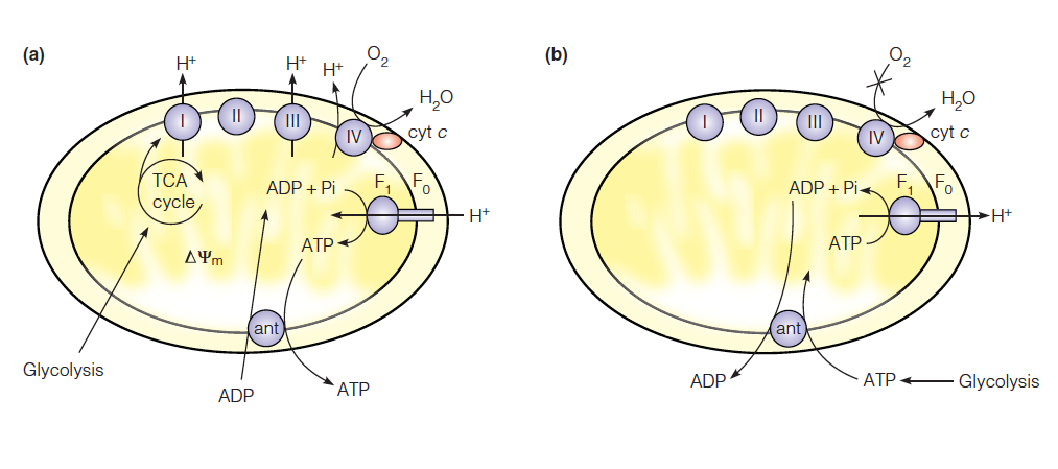
Reversibility of F1F0 ATP synthase
In normally respiring mitochondria, the pmf (proton motive force) is high and ATP synthase operates in the direction of ATP synthesis. The ATP is exchanged for cytoplasmic ADP via the adenine nucleotide translocase (ant).
During hypoxia(低氧), the pmf falls, and the cell relies mainly on homolactic (or alcoholic) fermentation for ATP production. This ATP enters the matrix in exchange for ADP.
- F1F0 ATP synthase operates as a proton-translocating ATPase, pumping protons out of the matrix, temporarily sustaining the pmf to support other processes, such as metabolite transport.
Major inner membrane transport systems for respiratory substrates and products
The ADP/ATP carrier and the phosphate translocase move substrates for oxidative phosphorylation (ADP and Pi) into the mitochondrion and the product (ATP) out.
Other transport systems move substrates and products for citric acid cycle oxidation into or out of the matrix, as dictated by the metabolic needs of the cell.
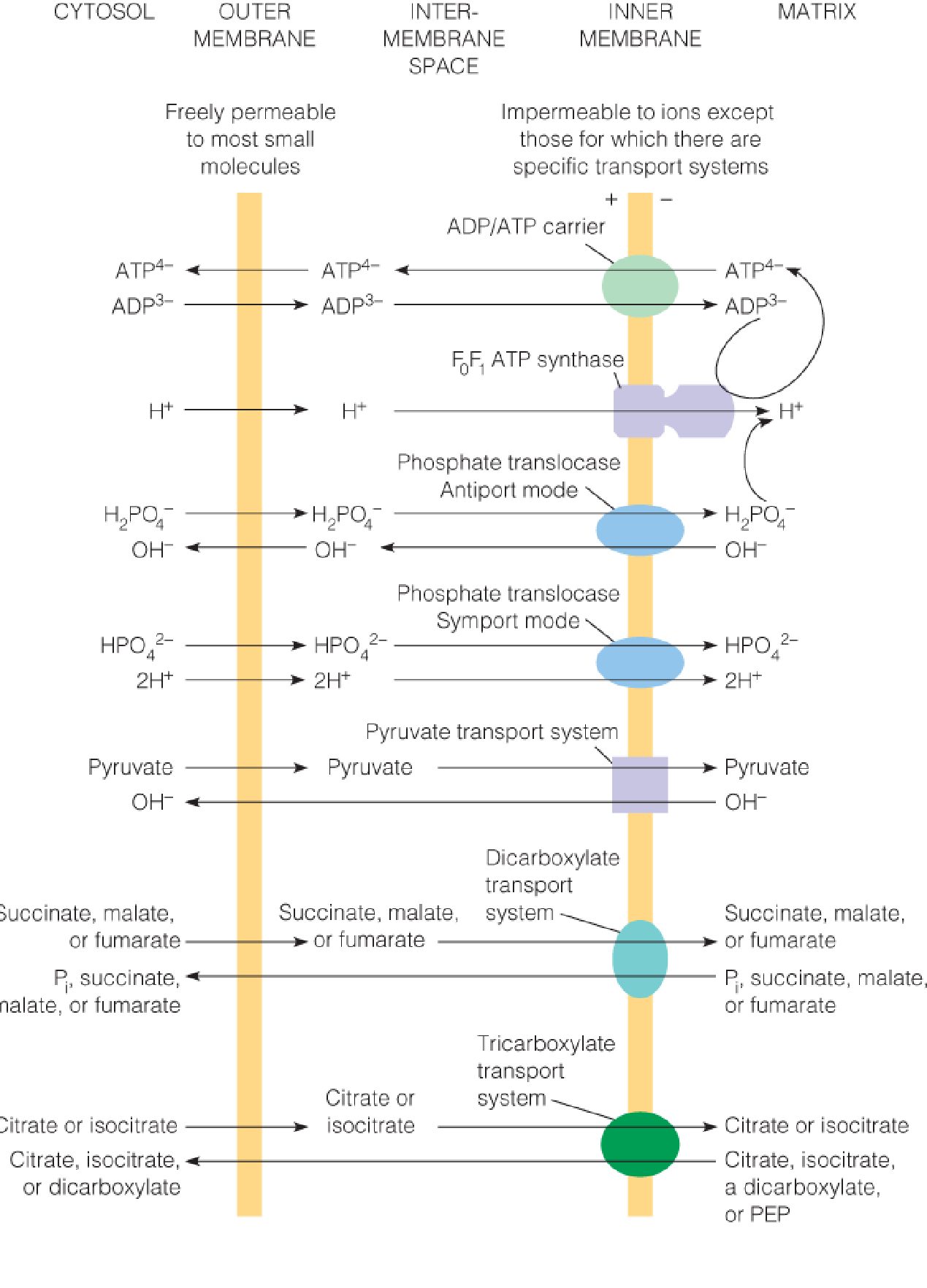
四、Mitochondrial Transport Systems
Electrons are transported into mitochondria by metabolic shuttles.
Reducing equivalents are shuttled from cytosol into mitochondria.
- The dihydroxyacetone phosphate/glycerol-3-phosphate shuttle.
- The malate/aspartate shuttle
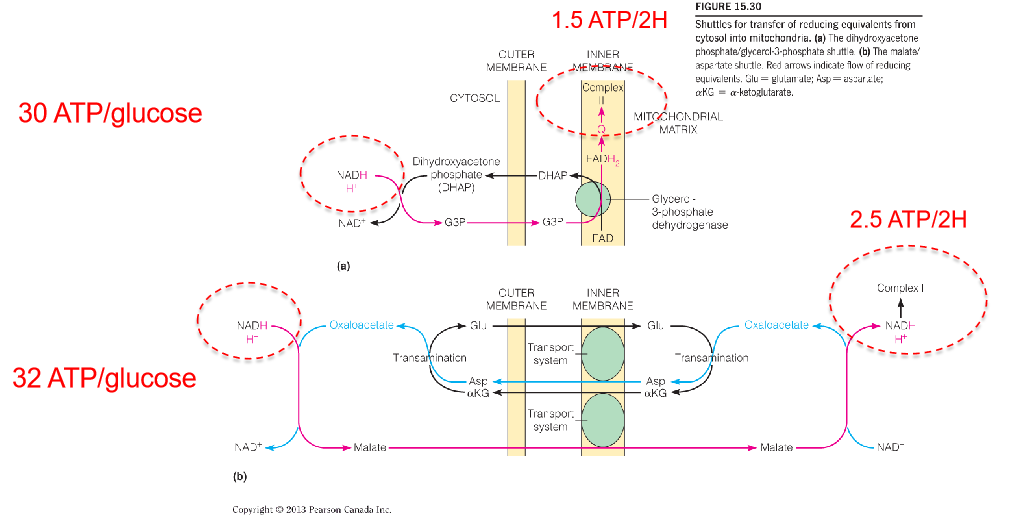
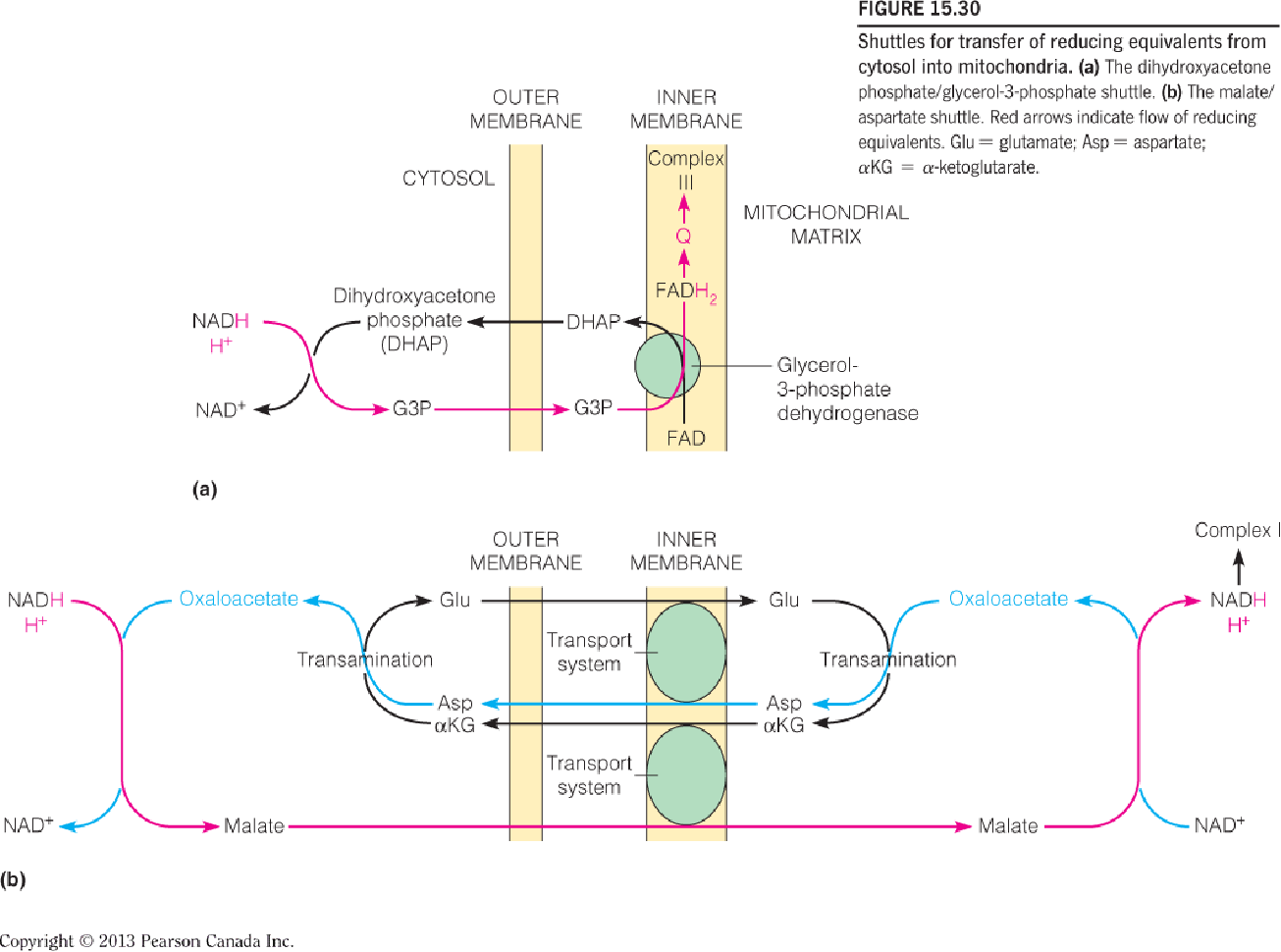
- The malate/aspartate shuttle:
- The OA is reduced into malate while the NADH is oxidized into NAD+.
- Through an antiporter, the malate is transferred into the mitochondrial matrix while an α-ketoglutarate is transferred out of the matrix.
- The NADH is reformed while the malate is re-oxidized into OA in the matrix.
- The OA is converted into aspartate while the glutamate transfer one amino group to OA and the glutamate molecule is converted into α-ketoglutarate which can be transported out of the matrix in step b).
- The aspartate is transported out of the matrix through another antiporter while a glutamate is transferred into the matrix.
- In the cytosol, the aspartate is converted into OA again through deamination and transfer one amino group to α-ketoglutarate and α-ketoglutarate is converted into glutamate.
- The dihydroxyacetone phosphate/glycerol-3-phosphate shuttle:
- The NADH and one proton deliver the hydrogen atom to DHAP molecule and forms G3P and NAD+.
- G3P is converted into OHAP in complex Ⅱ and the 2H atom is transferred into FAD and form FADH2.
- The 2H atom is transferred to CoQ(Q) and produce its reduced form QH2.
五、Energy Yields from Oxidative Metabolism
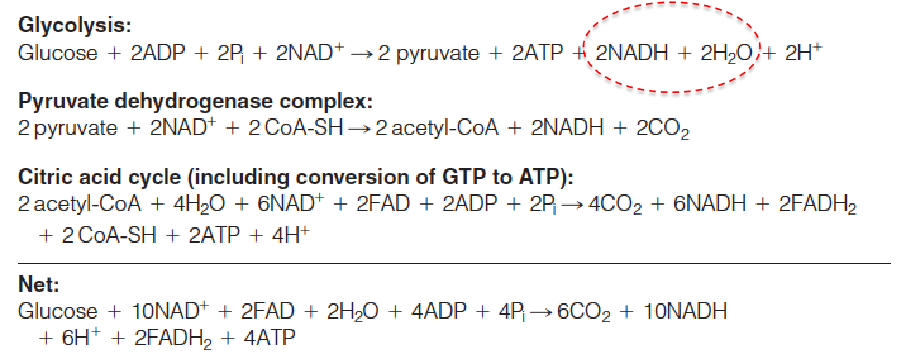
The complete oxidation of 1 mole of glucose generates about 30–32 moles of ATP synthesized from ADP.
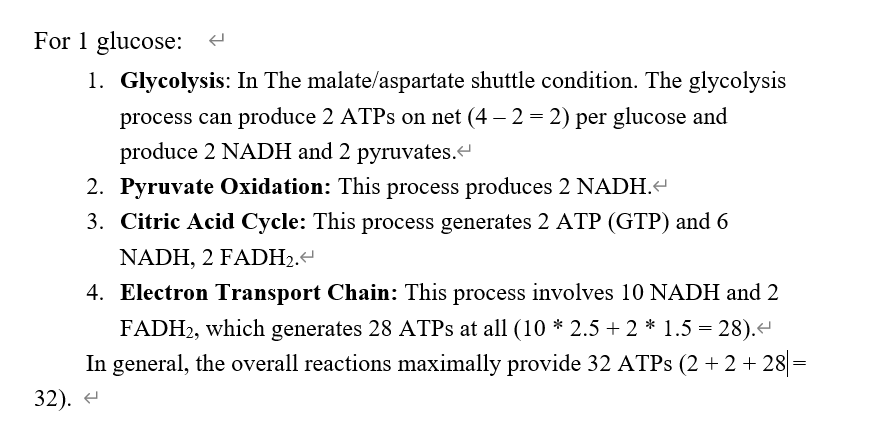
Now turn the NADH, FADH2, ATP all to ATP:
- In glycolysis, produces 5 or 7 ATPs, dur to the different shuttling method.
- In PDC: Produces 5 ATPs.
- In 3C cycle: produces 20 ATPs.
(32 x 30.5)/2870 = 34% under standard conditions
ATP hydrolysis is worth 50-60 kJ/mol in vivo
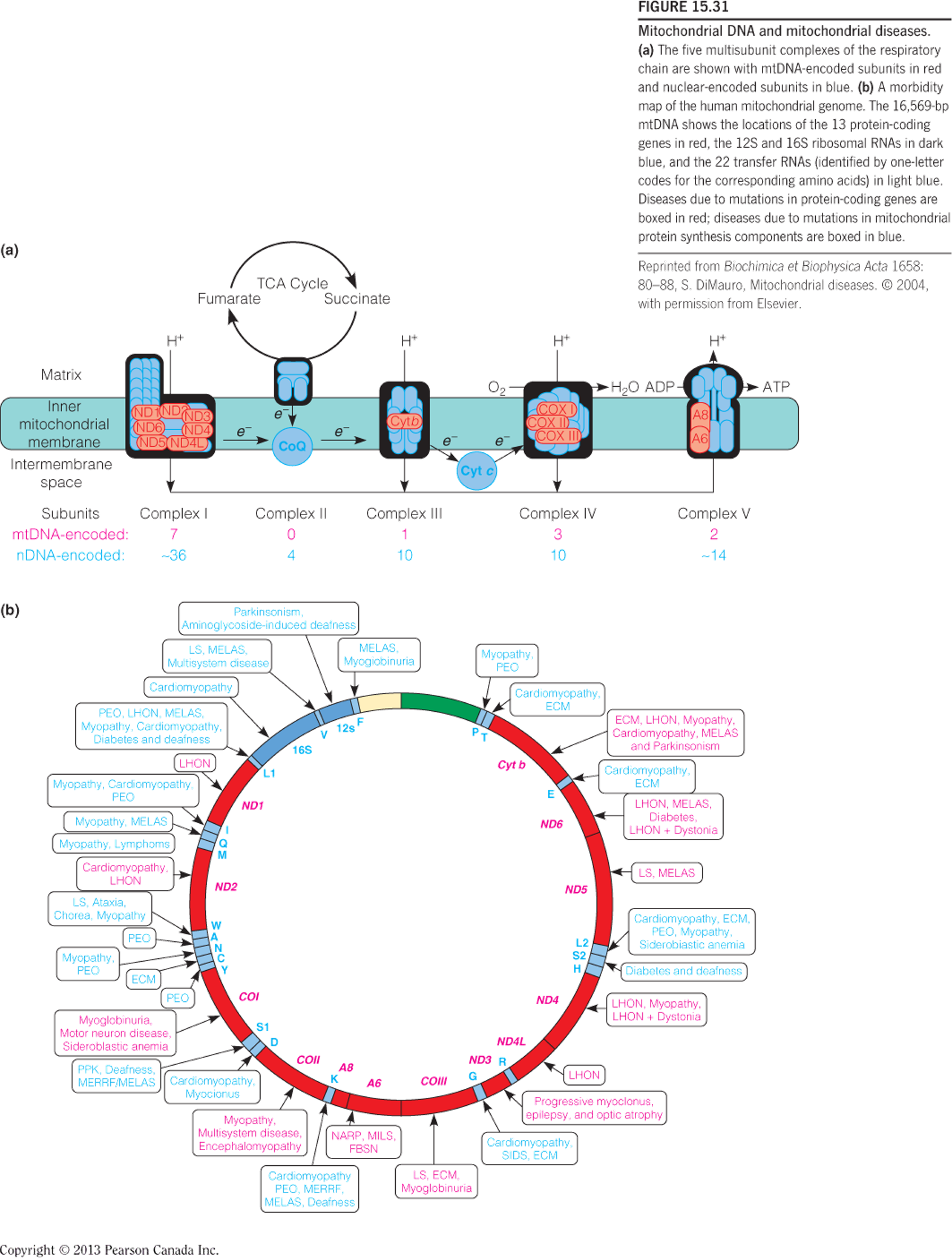
六、The Mitochondrial Genome and Disease
The five multisubunit complexes of the respiratory chain are shown with mtDNA-encoded subunits in red and nuclear-encoded subunits in blue.
A morbidity map of the human mitochondrial genome.
The 16,569-bp mtDNA shows the locations of the 13 protein-coding genes, the 12S and 16S ribosomal RNAs, and the 22 transfer RNAs.
Endosymbiont hypothesis (内共生假说)
七、Oxygen as a Substrate for Other Metabolic Reactions
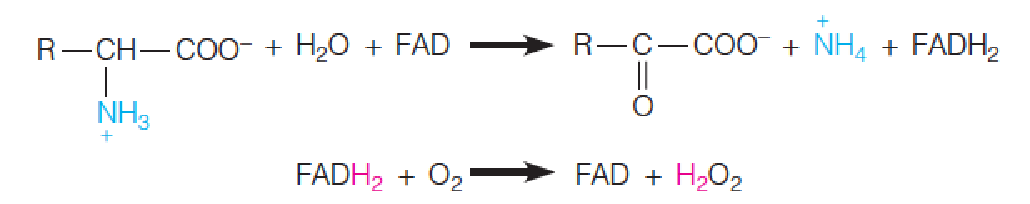
The term oxidase(氧化酶) is applied to enzymes that catalyze the oxidation of a substrate without incorporation of oxygen from O2 into the product.
A two-electron oxidation is usually involved, so the oxygen is converted to H2O2.
Most oxidases utilize either a metal or a flavin coenzyme.
D-Amino acid oxidases, for example, use FAD as a cofactor.
> 90% O
2is used by oxidative phosphorylation< 10% O2 by other metabolic reactions
200 enzymes use O2 as substrate
O2 is rather unreactive, so need metal ion or other cofactors
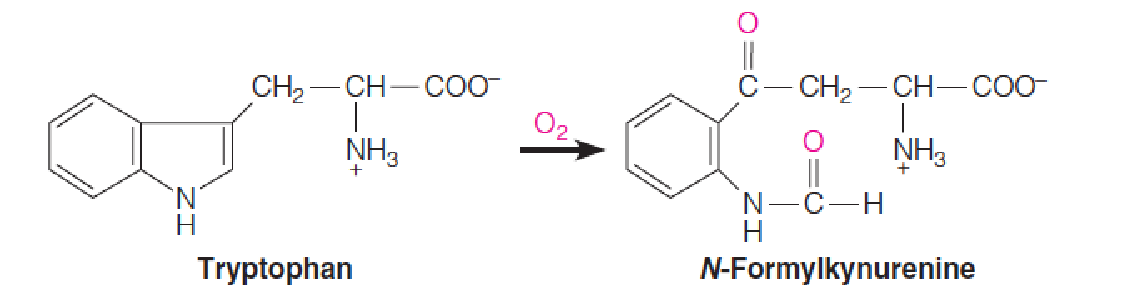
Oxygenases (加氧酶) are enzymes that incorporate oxygen atoms from O2 into the oxidized products; there are two classes—monooxygenases (单加氧酶) and dioxygenases (双加氧酶).
Dioxygenases, which incorporate both atoms of O2 into one substrate, are of limited distribution.
An example is tryptophan 2,3-dioxygenase, which contains a heme cofactor and catalyzes the first reaction in tryptophan catabolism:
Far more widely distributed are monooxygenases (单加氧酶), which incorporate one atom from O2 into a product and reduce the other atom to water.
A monooxygenase has one substrate (AH) that accepts oxygen and another (BH2) that furnishes the two H atoms that reduce the other oxygen to water.
Because two substrates are oxidized, enzymes of this class are also called mixed-function oxidases (混合功能氧化酶).
The general reaction catalyzed by monooxygenases is as follows:

Because the substrate AH usually becomes hydroxylated by this class of enzymes, the term hydroxylase (羟化酶) is also used.
An example of this type of reaction is the hydroxylation of steroids. Here the reductive cofactor, NADPH, appears as BH2.

Cytochrome P450
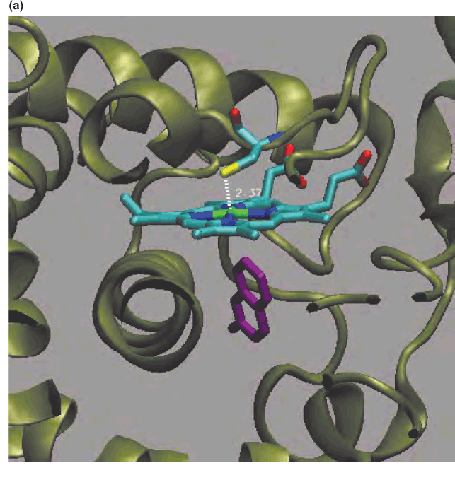
Active site of human cytochrome CYP2A6 in complex with the anticoagulant coumarin (香豆素).
The flavoprotein enzyme P450 oxidoreductase (POR) delivers electrons one at a time from NADPH to cytochrome P450.
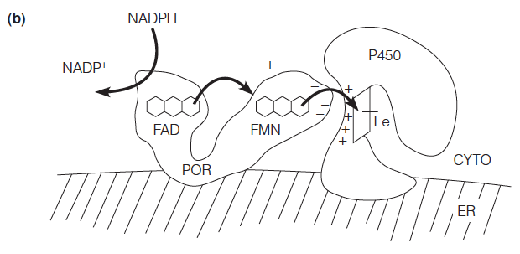
- Absorbs light strongly at 450 nm, heme
- Found in nearly all organisms
- Human genome has 57 genes for P450, a large and diverse protein family
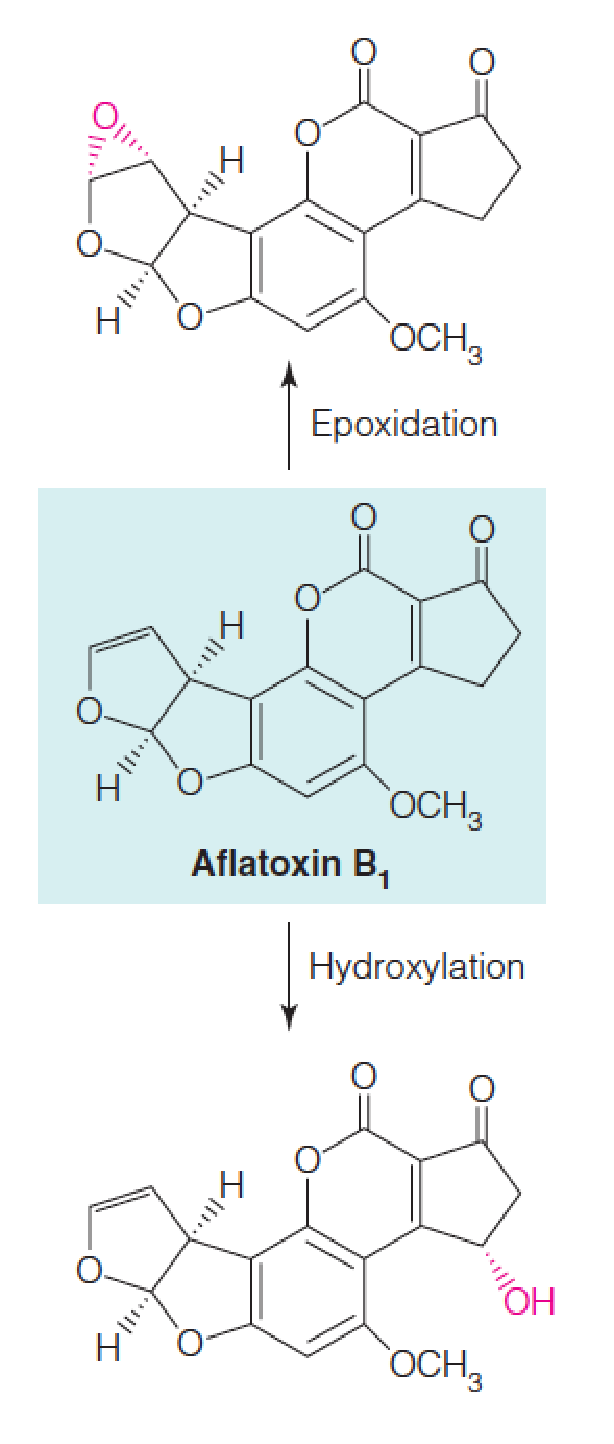
Cytochromes P450 are involved in hydroxylating a large variety of compounds.
These reactions include the hydroxylations in steroid hormone biosynthesis and the synthesis of hydroxylated fatty acids and fatty acid epoxides (环氧化合物).
In addition, cytochromes P450 act upon thousands of xenobiotics (外源化合物).
Hydroxylation of foreign substances usually increases their solubility and is an important step in their detoxification, or metabolism and excretion.
However, some of these reactions result in activation of potentially carcinogenic substances to more reactive species, as shown for aflatoxin (黄曲霉毒素) B1 which is converted to more reactive species either by hydroxylation or epoxidation.
八、Reactive Oxygen Species, Antioxidant Defenses, and Human Disease
Reactive oxygen species (ROS, 活性氧簇)
The generation and interconversion of the most common reactive oxygen species are shown:
Superoxide 过氧化物
Hydroxyl radical 羟自由基
Nitric oxide 一氧化氮
Peroxynitrite 过氧亚硝酸盐
Oxidized coenzyme Q (氧化辅酶Q)
Reactive oxygen species (ROS,活性氧类) are chemically reactive chemical species containing oxygen. Examples include peroxides, superoxide, hydroxyl radical, singlet oxygen(单线态氧), and alpha-oxygen.
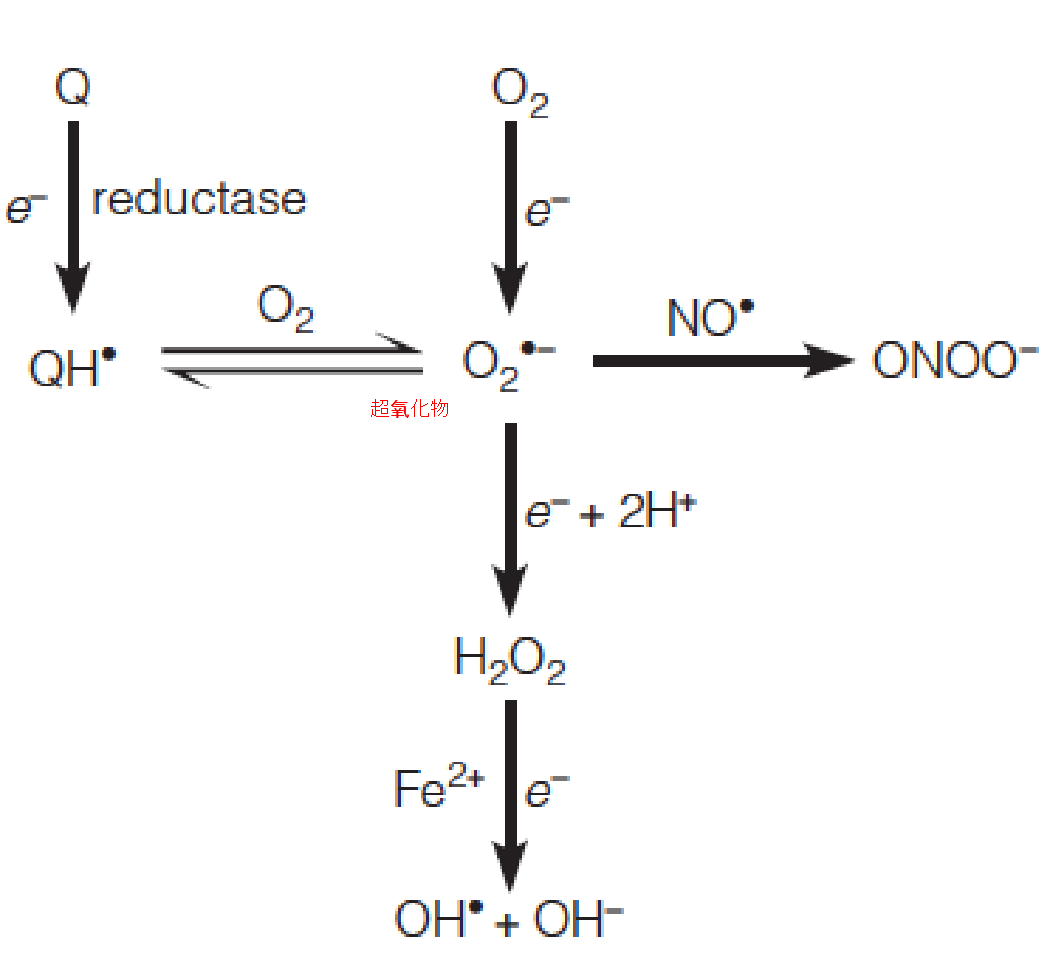
Incompletely reduced oxygen species
Unpaired electron
Mitochondrion is a primary source of ROS
1. Hydroxyl Radical
Hydroxyl radical is the most active mutagen derived from ionizing radiation.
Hydroxyl radical is also produced from H2O2 in the Fenton reaction:
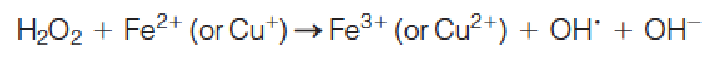
2. Superoxide
Superoxide per se is relatively nontoxic.
Because it contains an unpaired electron, it is a free radical, and it combines readily with another free radical, nitric oxide (NO, 信号分子,促进血管扩张), a biological signaling agent that is produced in many animal tissues.
The product is peroxynitrite also considered a reactive oxygen species.
Peroxynitrite causes lipid peroxidation and also causes nitration of tyrosyl hydroxyl groups in proteins, a reaction particularly damaging to membrane proteins.
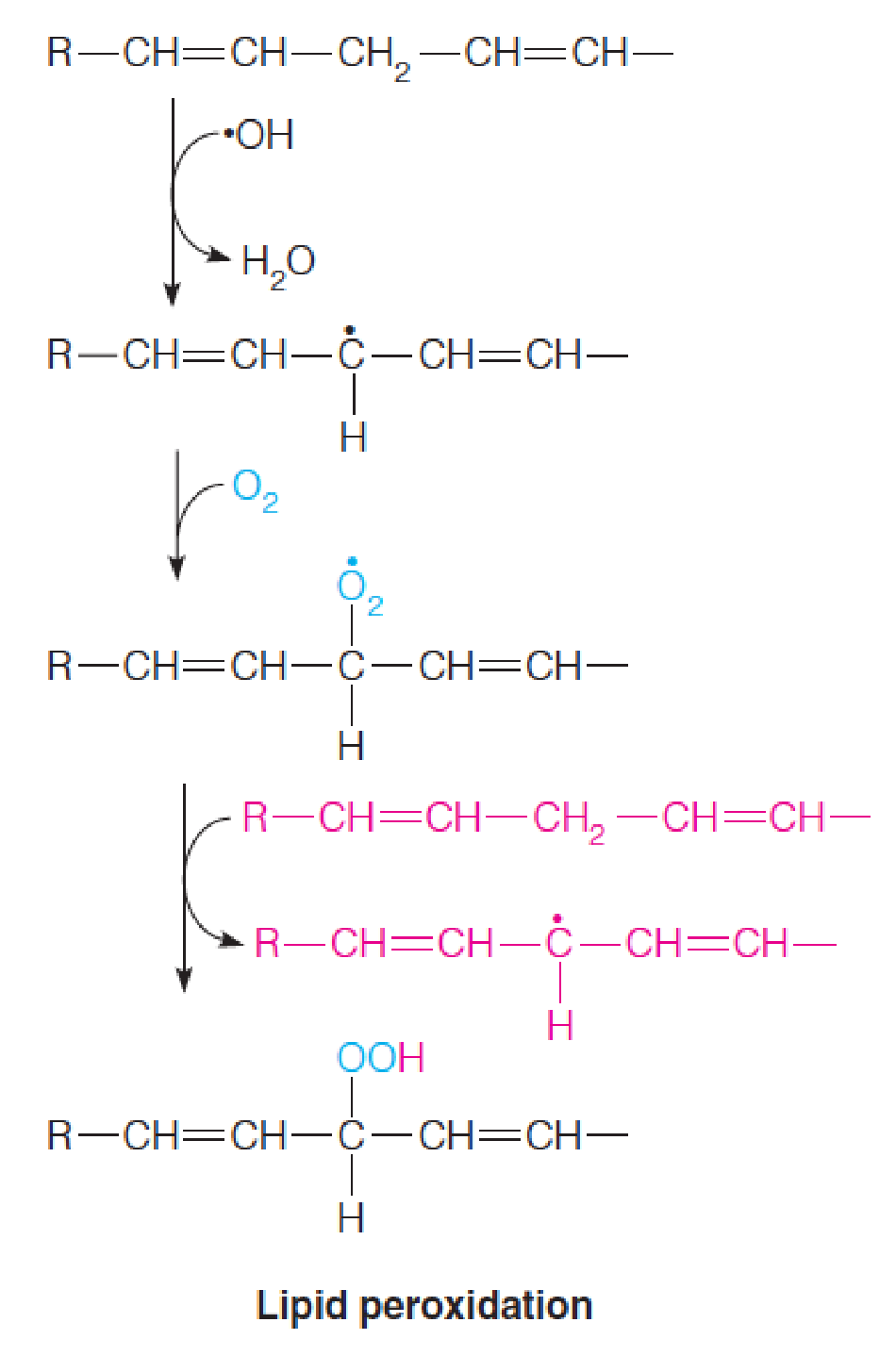
Hydroxyl radical also damages nucleic acids, both by causing polynucleotide strand breakage (double-stranded DNA breaks are lethal) and by changing the structure of DNA bases.
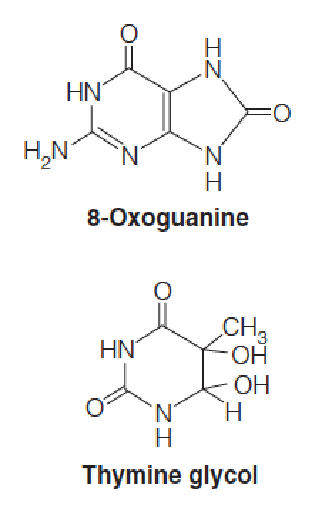
About 20 different base changes, or DNA lesions, are known to result from reactions of hydroxyl radical with DNA.
Some lesions are mutagenic because the altered base created, such as 8-oxoguanine (8-氧鸟嘌呤), forms non-Watson–Crick base pairs during DNA replication. (Cause DNA mutation)
Other lesions, such as thymine glycol (胸腺嘧啶二醇), are potentially lethal because, unless the lesion is repaired their occurrence in DNA blocks replication past that site.
Uncontrolled overproduction of reactive oxygen species has the potential to inflict considerable damage on the tissues in which they are produced, a situation called oxidative stress.
A series of elaborate mechanisms has evolved to minimize its harmful consequences.
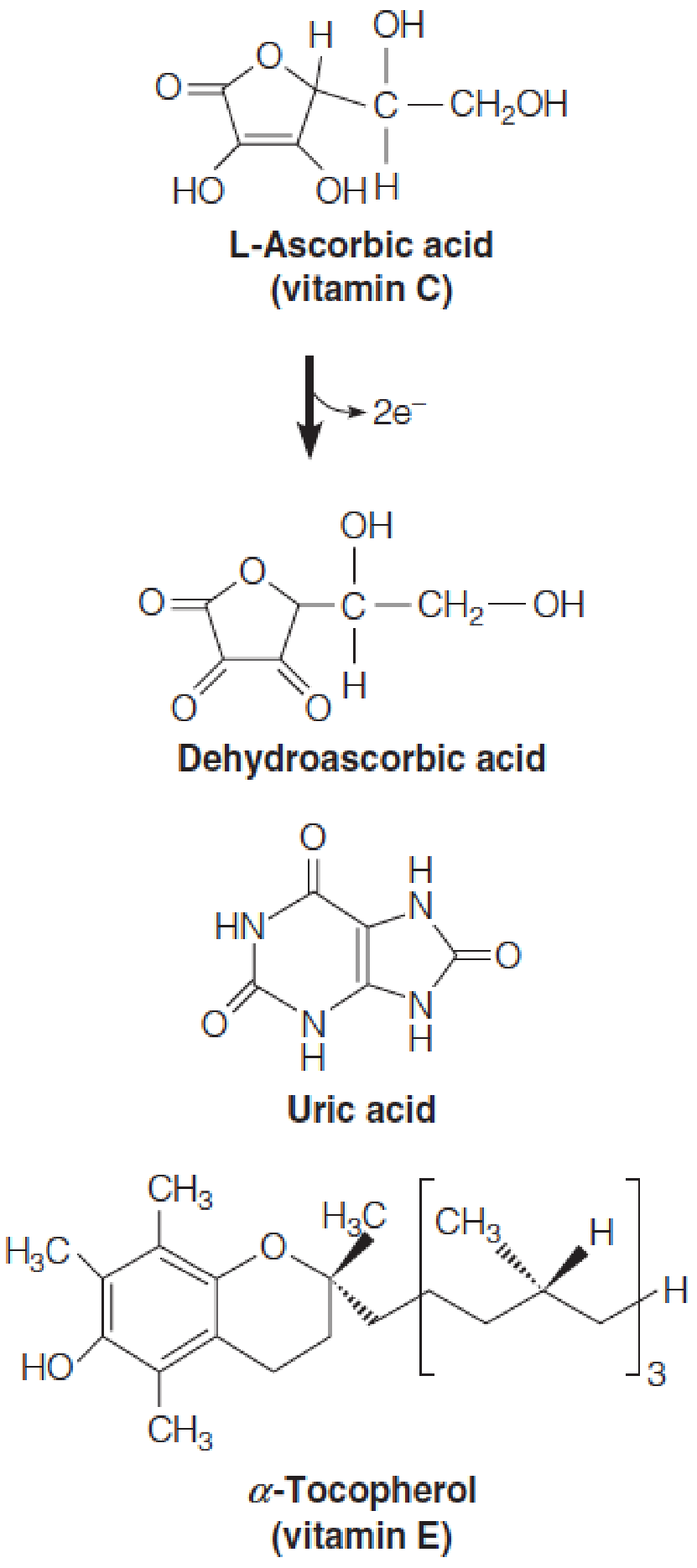
The nonenzymatic protection is afforded by antioxidant compounds:
- Glutathione 谷胱甘肽
- Vitamins C and E
- Uric acid 尿酸
These compounds can scavenge ROS before they can cause damage, or they can prevent oxidative damage from spreading.
- Blueberry reduces ROS
- Smoking increases ROS
Disease
Among enzymatic mechanisms the first line of defense is superoxide dismutase (SOD, 超氧化物歧化酶), a family of metalloenzymes that catalyze a dismutation (a reaction in which two identical substrate molecules have different fates).
Here, one molecule of superoxide is oxidized to H2O2 and one is reduced to O2.
Hydrogen peroxide (过氧化氢) is metabolized either by peroxiredoxins (过氧化还原酶), a ubiquitous family of thiol proteins, catalase (过氧化氢酶), another widely distributed enzyme, or by a more limited family of peroxidases (过氧化物酶).

Oxidative stress: damages lipids, proteins, and nucleic acids; causes cardiovascular disease, cancer, stroke, neurodegenerative diseases, chronic inflammatory diseases, aging
Amyotrophic lateral sclerosis (ALS): 肌萎缩侧索硬化; AKA motor neuron disease (MND) or Lou Gehrig’s disease
Catalase (过氧化氢酶) is a heme protein with an extremely high turnover rate (> 40,000 molecules per second).
It catalyzes the following reaction:
2 H2O2 $\rarr$ 2 H2O + O2
Peroxidases (过氧化物酶), which are widely distributed in plants, reduce H2O2 to water at the expense of oxidation of an organic substrate.
An example of a peroxidase is found in erythrocytes, which are especially sensitive to peroxide accumulation.
Within erythrocytes is glutathione peroxidase, an enzyme that reduces H2O2 to water, along with the oxidation of glutathione.
2 GSH + H2O2 $\rarr$GSSG + 2 H2O