L03 RNA and Protein
一、RNA
Classes of RNA Molecules
Major classes, all involved in protein synthesis, and all of which undergo RNA processing after transcription (“post-transcriptional modifications”).
messenger RNA (mRNA) – encodes amino acids in codons of 3 nucleotides.
- Polycistronic mRNAs found in bacteria
- untranslated regions at 5’ and 3’ ends that may contain regulatory sequences.
ribosomal RNA (rRNA) – part of the ribosome that produces proteins.
transfer RNA (tRNA) – align amino acids to mRNAs during translation; reads three nucleotides of mRNA and converts to one amino acid.
Small nuclear RNA (snRNA) is involved in mRNA splicing.
Small nucleolar RNA (snoRNA) is involved in rRNA processing.
RNA Production
RNA is produced from DNA via Transcription
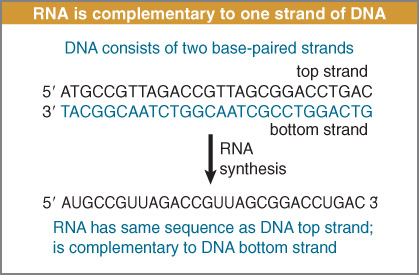
RNA is synthesized by using one strand of DNA as a template for complementary base pairing.
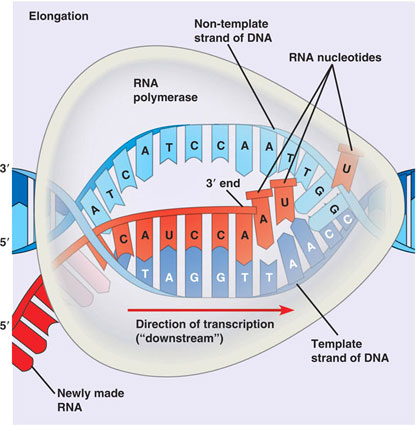
The gene may be longer than the sequence coding for protein.
In eukaryotes, transcription occurs in the nucleus, and translation occurs in the cytoplasm.
In contrast, transcription and translation take place in the same compartment in bacteria.
Components of RNA
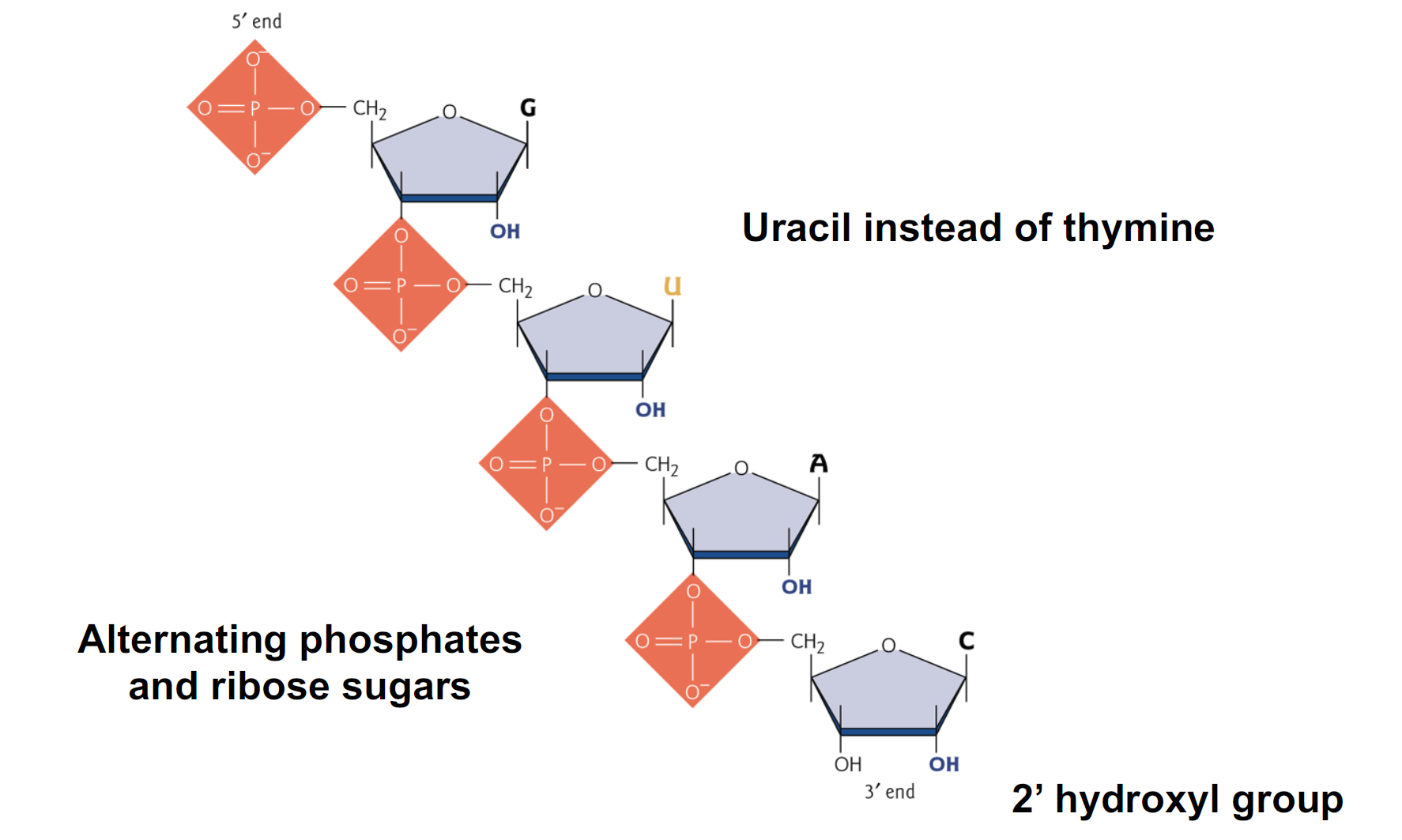
- Uracil instead of thymine
- Alternating phosphates and ribose sugars
- 2’ hydroxyl group
RNA Structures
1. RNA Secondary Structure
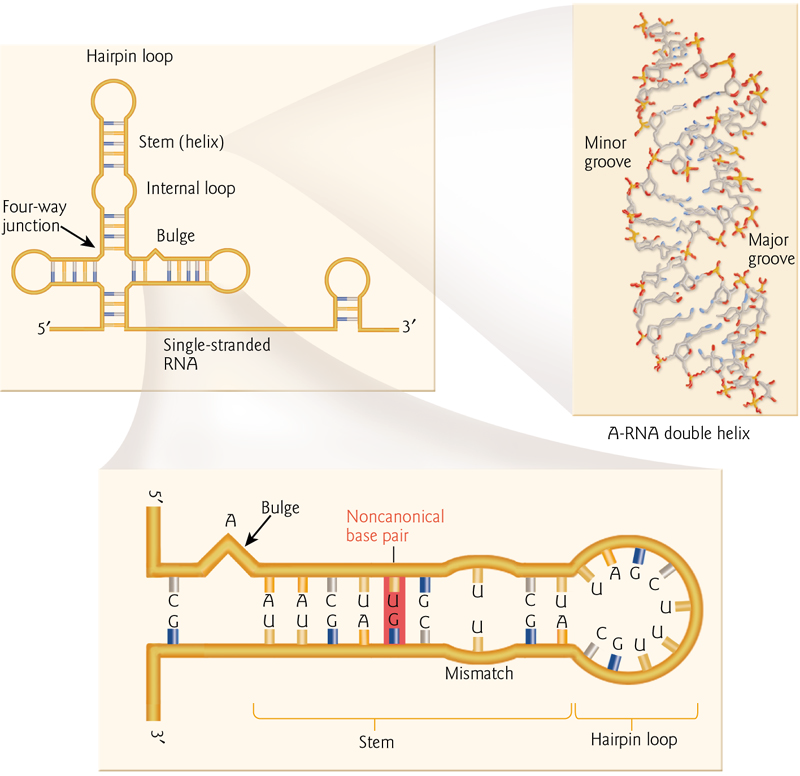
Typical structures formed by folded RNA include non-canonical base pairs (“wobble”)
hairpins
loops
bulges
junctions
RNA can also form double helices
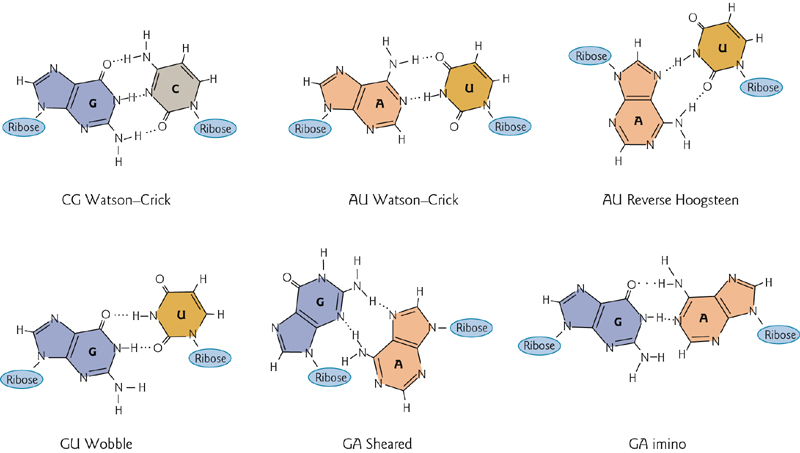
2. Base Pairing in Double Strand RNA
R—R $>$R—D>D—D
In addition to “normal” (Watson-Crick pairing), there are four noncanonical pairs that can form by hydrogen bonding.
3. Base Pairing in Triple Strand RNA
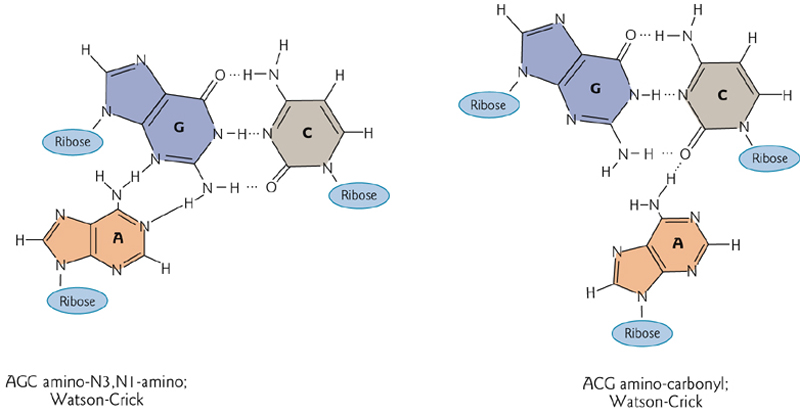
Hydrogen bonding stabilizes a triplex of three nucleotides.
4. Secondary And Tertiary tRNA Structures
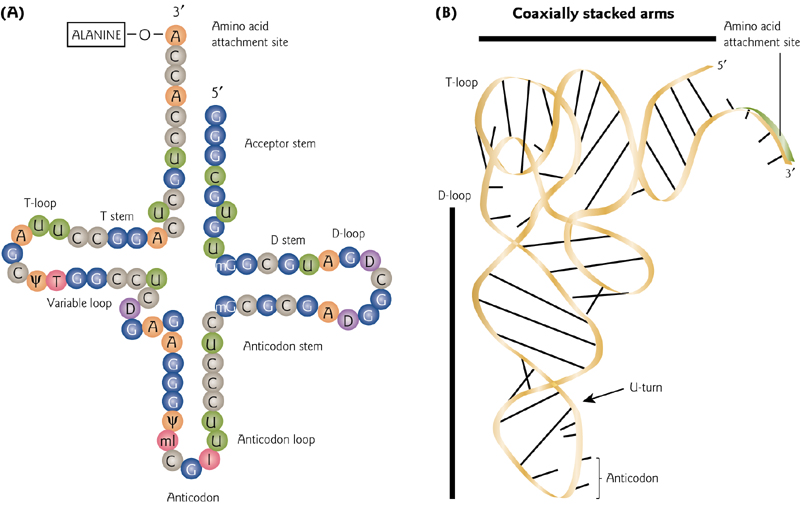
Typical “cloverleaf” drawing versus a more realistic 3-D tRNA structure. The “anticodon” is the functionally important bit, and it sticks out to remain accessible.
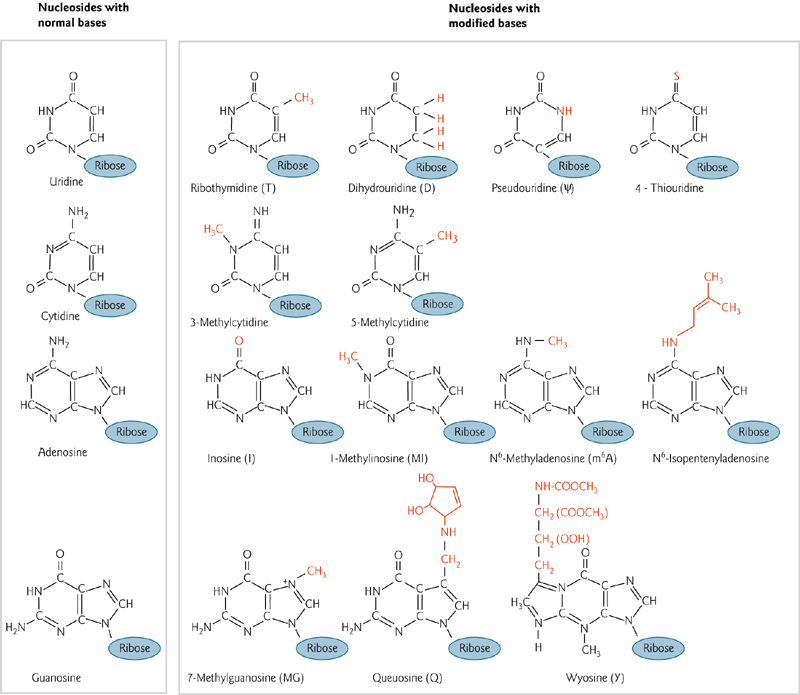
5. Modified Bases
More than 50 modified bases are found in RNA
6. RNA Pseudoknots
RNA pseudoknots comprise functional domains within ribozymes, self-splicing introns, ribonucleoprotein(核蛋白,RNP) complexes, viral genomes, and other biological systems.
An important piece of RNA architecture for the globular structure capable of performing important biological functions.
First found in plant viruses
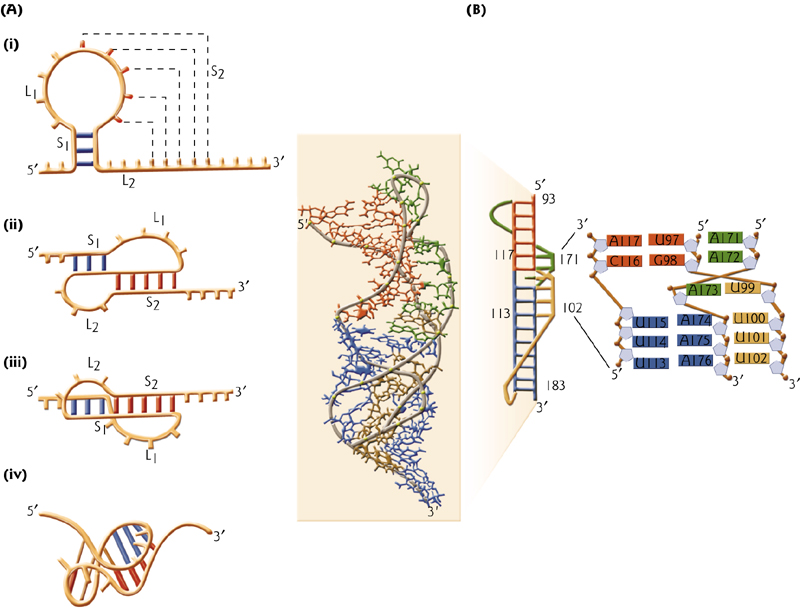
假结结构最初于1982年在芜青花叶病毒中发现 (First found in plants)。假结结构折叠成类似于绳结的三维结构,但却并不是真正的拓扑学的结。
7. structural motif 结构模体
结构模体(英语:structural motif,亦称为结构基序)是链状生物分子(如蛋白质或核酸)中的一种超二级结构,也存在于其它分子之中。结构模体使得我们无法预测蛋白的生物学功能:存在于蛋白质与酶中的模体可能功能迥异。
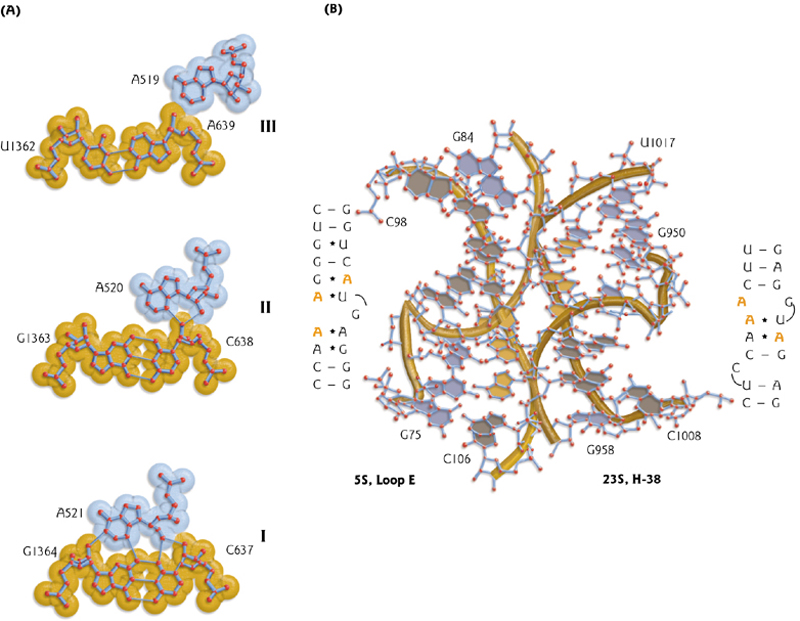
A-minor motif
A common “long range” interaction among different parts of the RNA molecule. Good fit between adenosine(腺苷) and the minor groove of a RNA double helix.
(RNA分子的不同部分之间的一种常见的长期相互作用。在腺甘酸和RNA双螺旋的minor groove之间有很好的匹配。)
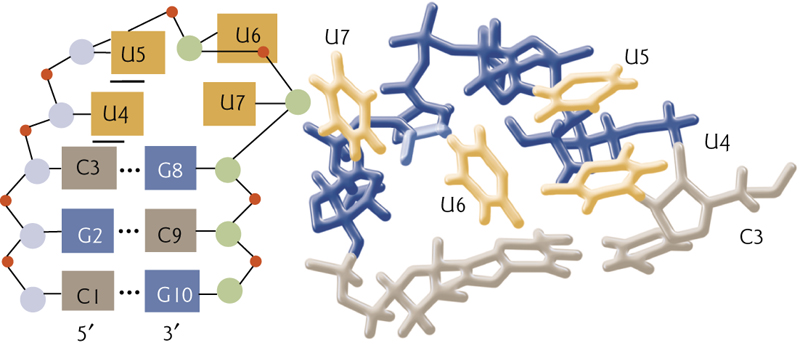
Tetraloop motif
In tetraloop motif, base breaking interactions stabilize the structure
在Allison的文章中描述的其他结构包括ribose zipper motif和kink-turn motif,两者都与其他结构结合,定义了广泛的RNA结构。
8. Protein-Mediated RNA Folding
Unfolded RNA can become misfolded, or can directly fold into the proper structure.
Binding proteins (blue) can stabilize the structure.
RNA “chaperones(分子伴侣)” can prevent misfolding.
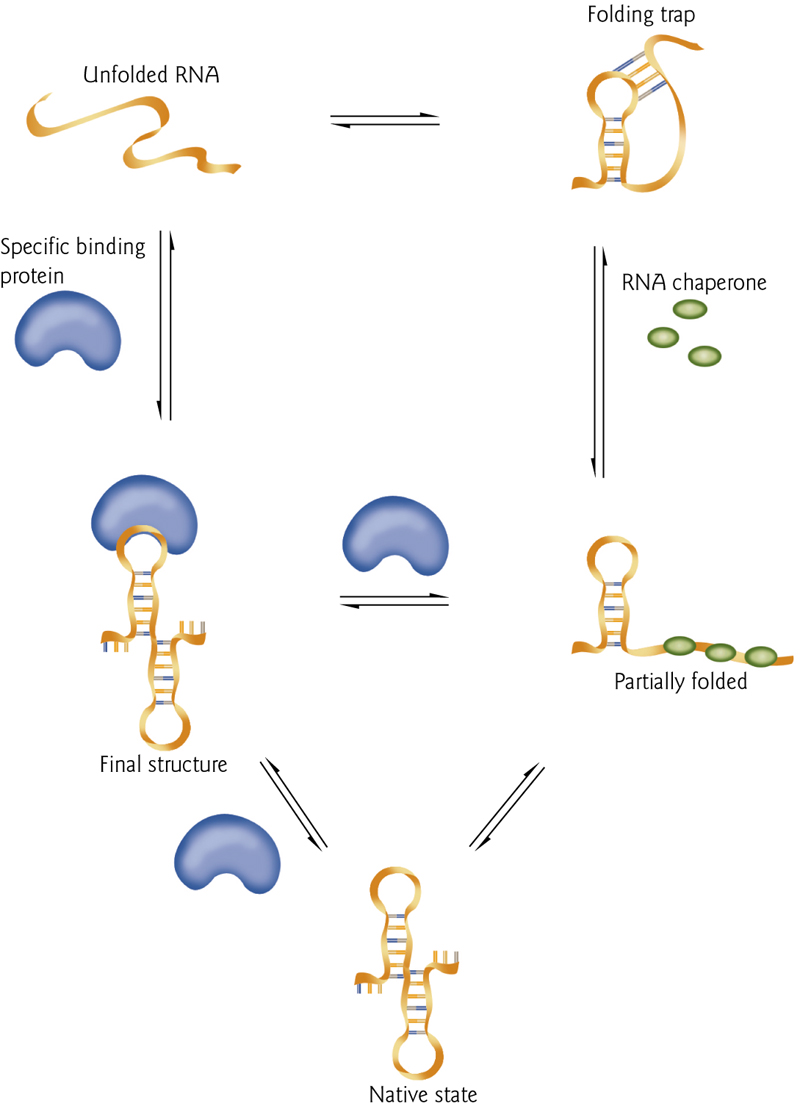
9. RNA Structure can be assayed by Cleavage of The Backbone
RNA结构可通过主链的裂解来测定
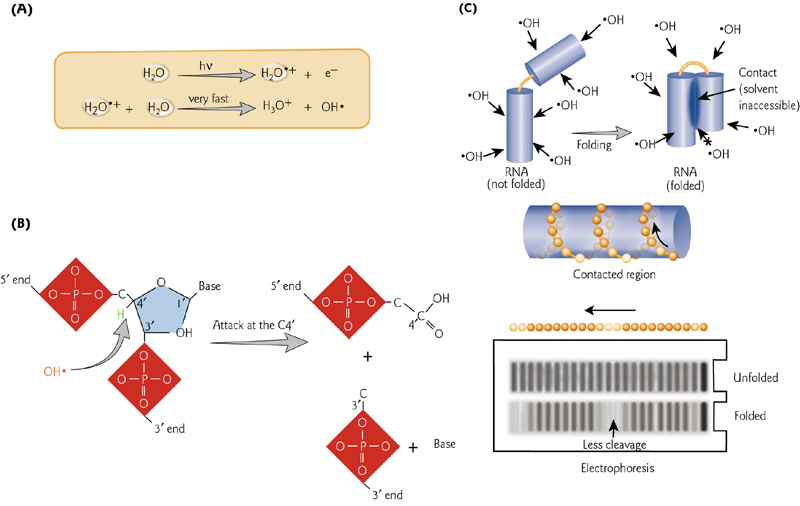
Hydroxyl radicals cleave at unprotected nucleotides. In a folded structure, specific nucleotides are protected from cleavage.
The RNA World
This theory indicates the evolution starts at RNA
RNA can perform many of the functions needed by a cell, including encoding information, performing functions as catalysts (“ribozymes”), regulating gene expression, binding proteins, etc.
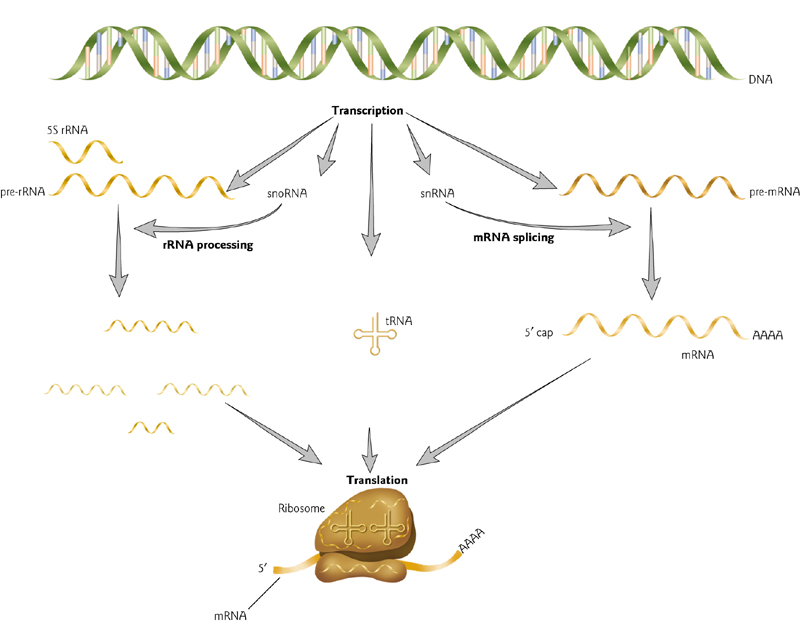
5 Major Classes of RNAs During Gene Expression
- snoRNA
- tRNA
- snRNA
- mRNA
- rRNA
Ribozyme 核酶
1. Self-Splicing
Thomas Cech demonstrated splicing of the “R loop” (or intervening sequence, IVS – an “intron”) in the absence of protein.
(托马斯·切赫证明了在缺乏蛋白质的情况下R环(或插入序列,IVS内含子)的剪接。)
2. Ribozyme can catalyze reactions - like proteins
The maturation of a tRNA is catalyzed by RNase P (with protein using RNA as a co-factor).
The maturation of a tRNA (production of 3’ and 5’ products) requires protein (Rpp) and RNA.
Hammerhead ribozymes – A frequent catalytic motif of plant viroids
Viroids = pathogenic RNAs.
The catalytic motifs of these small ribozymes are all involved in replication via “rolling circles”.
Long RNAs are self-cleaved into monomers.
Small ribozymes of 40 to 154 nt
May date from the “RNA world”
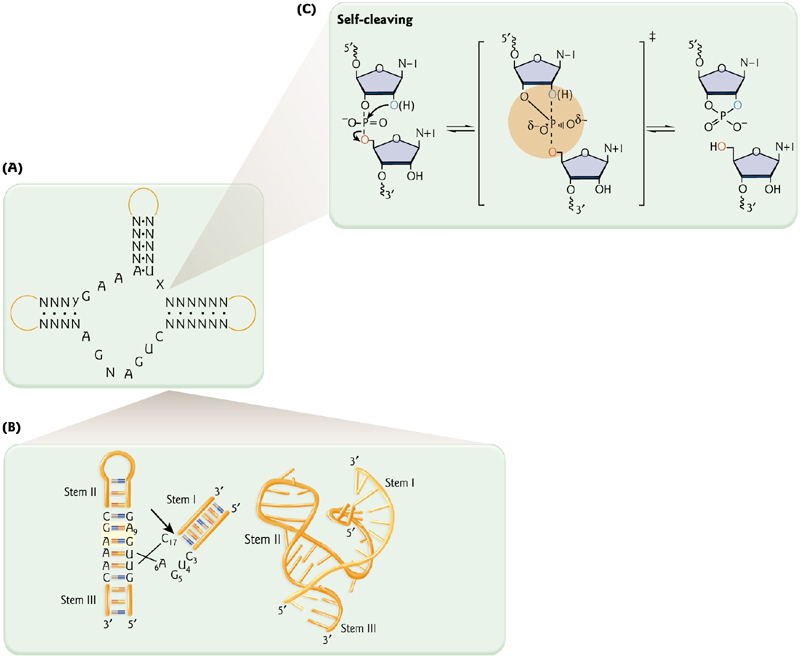
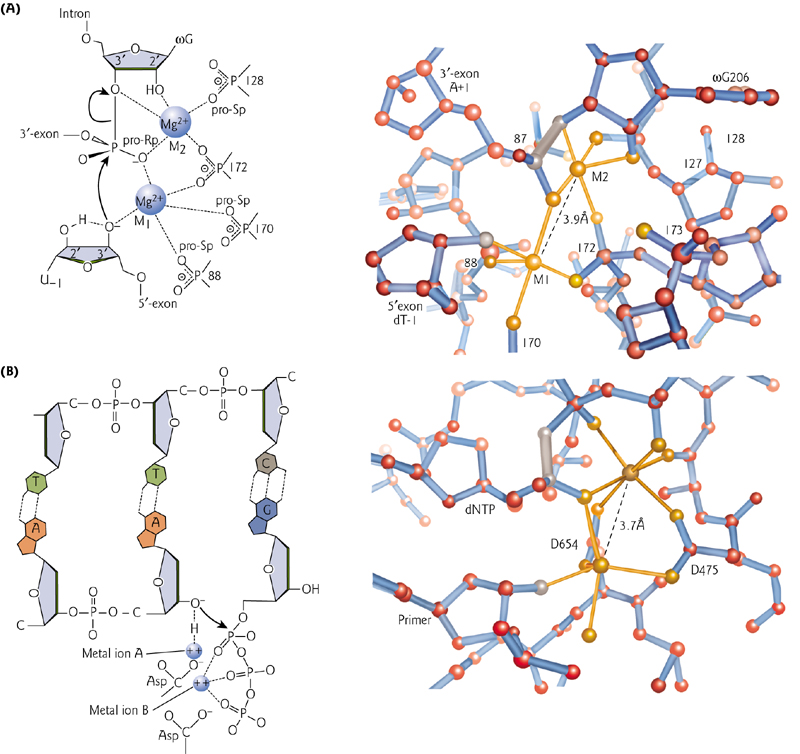
3. Structural similarities between RNA ribozyme and protein Polymerase
Both a self-splicing intron and bacteriophage T7 DNA polymerase contain two metal ions that coordinate the substrate position
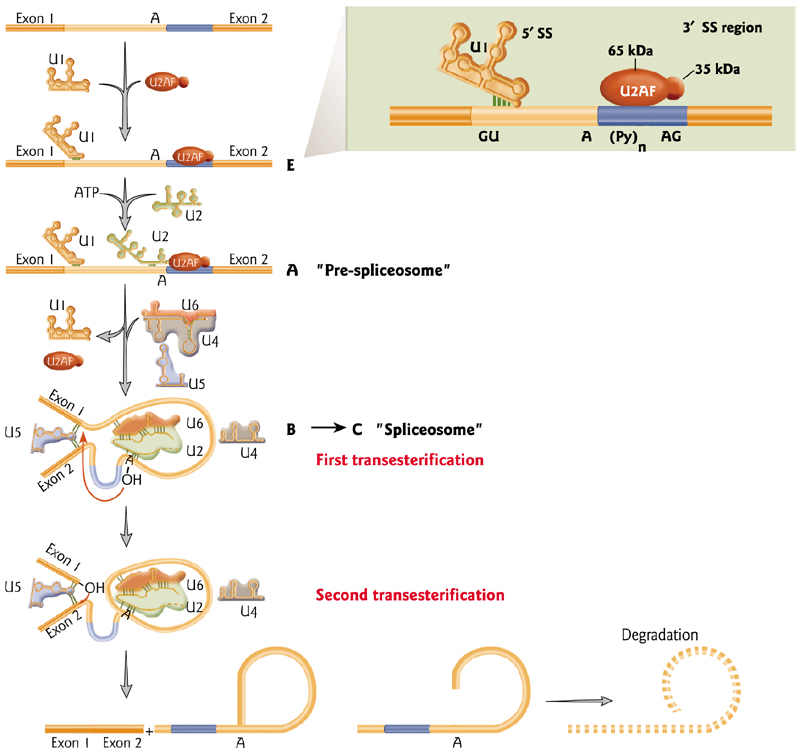
二、Post-transcriptional Events
Mature mRNA is shorter than the gene that encode it
- Evidence to suggest that portions of the mRNA (introns) are removed in a post-transcriptional processing event
Splicing
Splicing is the mechanism that removes introns
Signals in the mRNA determine which portions are removed from the mRNA to produce the mature product.
Splicing is more prevalent(普遍的) in higher eukaryotes
The introns size can varies from short to very long
Related genes diverge in the introns
- The sequences of the mouse amaj- and amin- globin genes are closely related in coding regions but differ in the flanking regions and long intron.
Splicing requires specific riboproteins that produce a transesterification reaction to remove the intron via production of a “Lariat” structure
Transesterification: 转酯(作用),酯基转移(作用),酯交换(作用)
Lariat: 套索
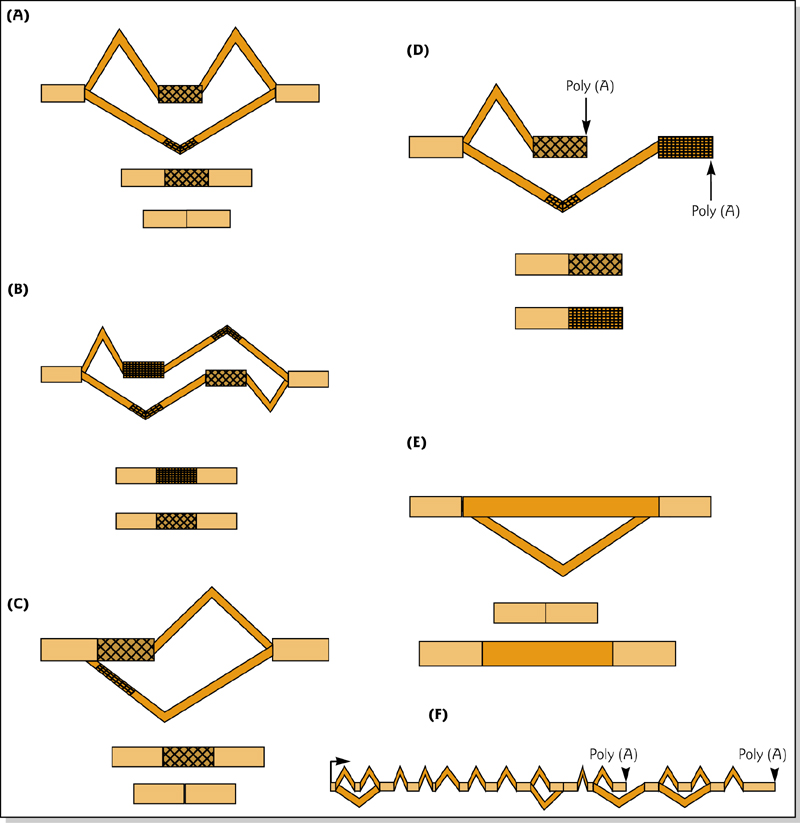
1. Alternative Splicing
One gene can produce a huge number of mRNA variants. This impacts protein diversity, localization, stability as well as mRNA turnover.
Alternative splicing is a tremendous source of variation in protein sequences
mRNA Maturation
The mRNA Maturation process involves many modification steps
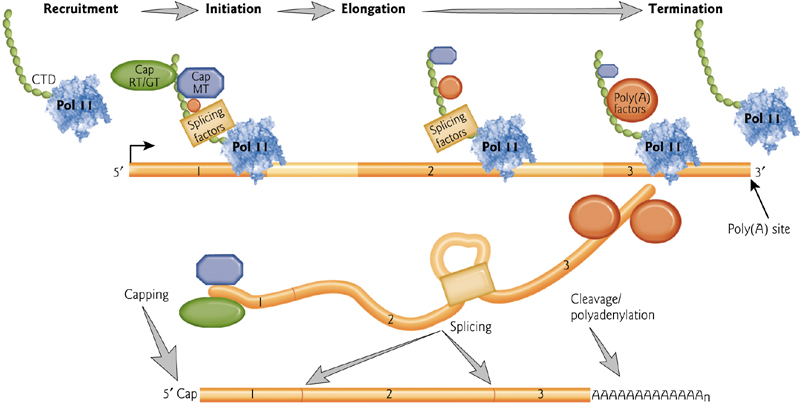
1. 5’ cap and a 3’ poly-A tail
In addition to splicing and removal of introns, a 5’ cap and a 3’ poly-A tail are added to protect the mRNA from degradation.
5’ mRNA Gap
The gap contains an unusual 5’-5’ bond
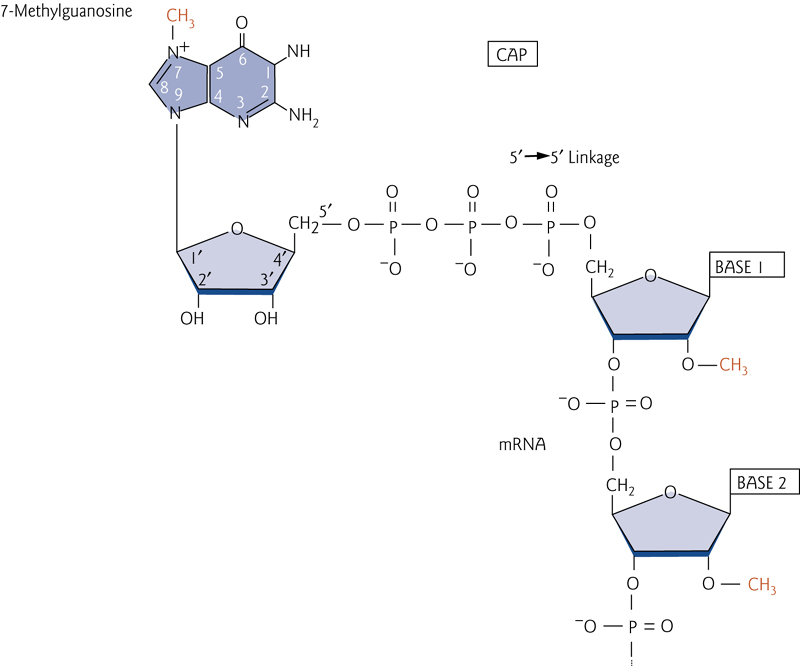
A 7-methylguanosine nucleotide is attached via a 5’-5’ linkage to the 5’ end of the mRNA, protecting it from degradation from exonucleases.
3’ mRNA poly tail
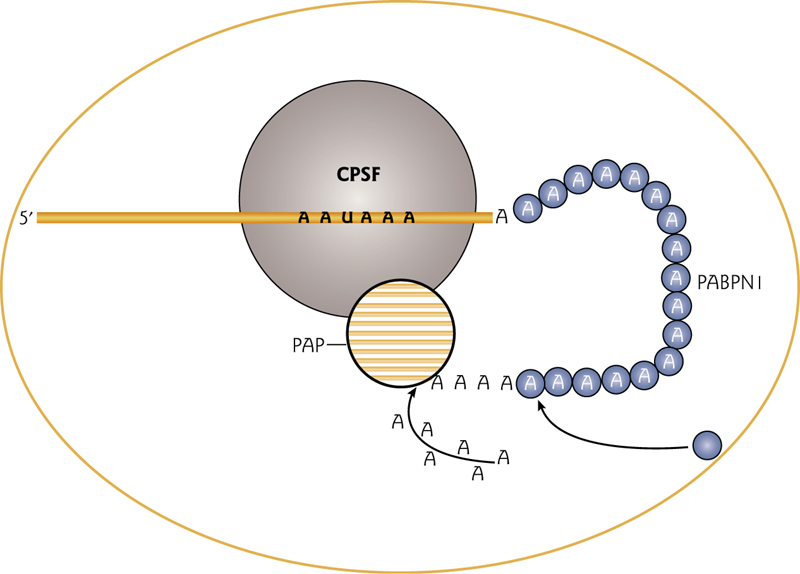
The cleavage and polyadenylation specificity factor (CPSF) binds to the polyadenylation signal AAUAAA at the 3’ end of the mRNA.
The Poly(A) binding protein PABPN1 binds the growing poly(A) tail.
These proteins recruit and increase processivity of the poly(A) polymerase
2. mRNA can be changes by a type of editing after production
This is evident by comparison of a cDNA product to the DNA of the gene that encodes it.
Rather than splice variants, sometimes small numbers of nucleotides have been added or removed.
This happens most commonly in the mitochondria
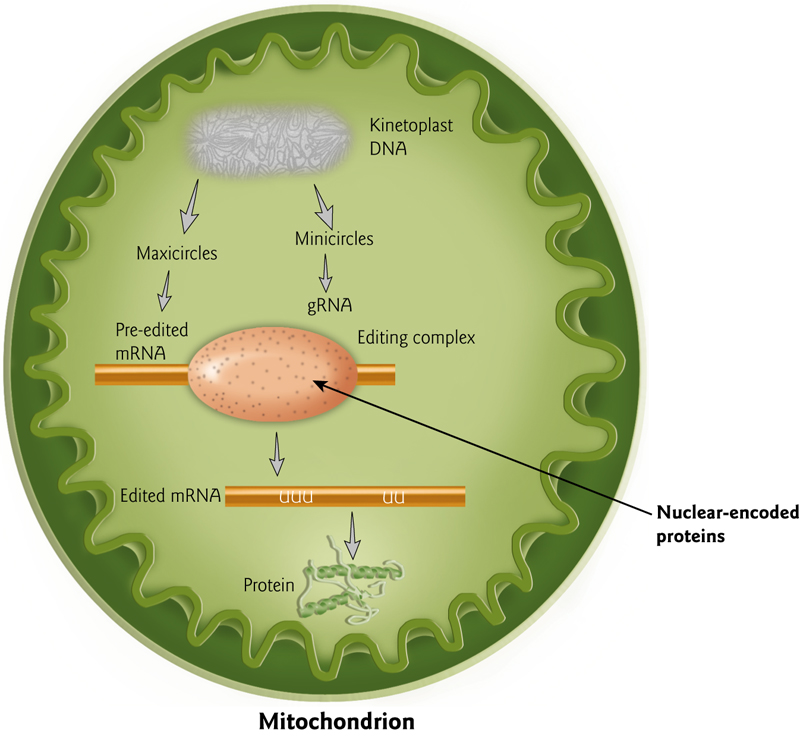
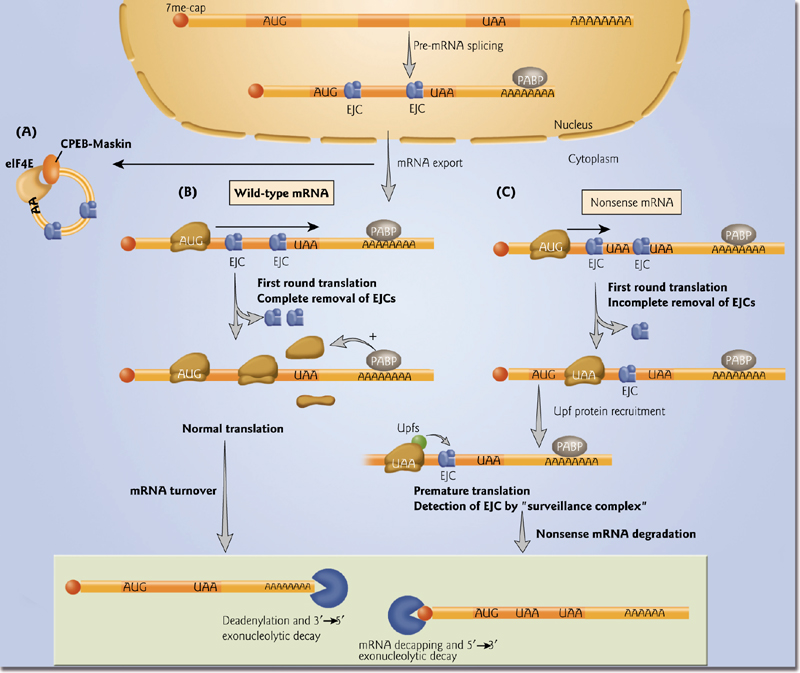
3. Check the quality of translation
- Some ribonucleo proteins (RNPs) are stored in the cytoplasm in an inactive state.
- Normal translation –ribosomes displace exon-exon junction complexes (EJCs) from mRNAs. General mRNA decay pathways.
- Nonsense-mediated decay results when EJCs are not removed
三、Gene to proteins
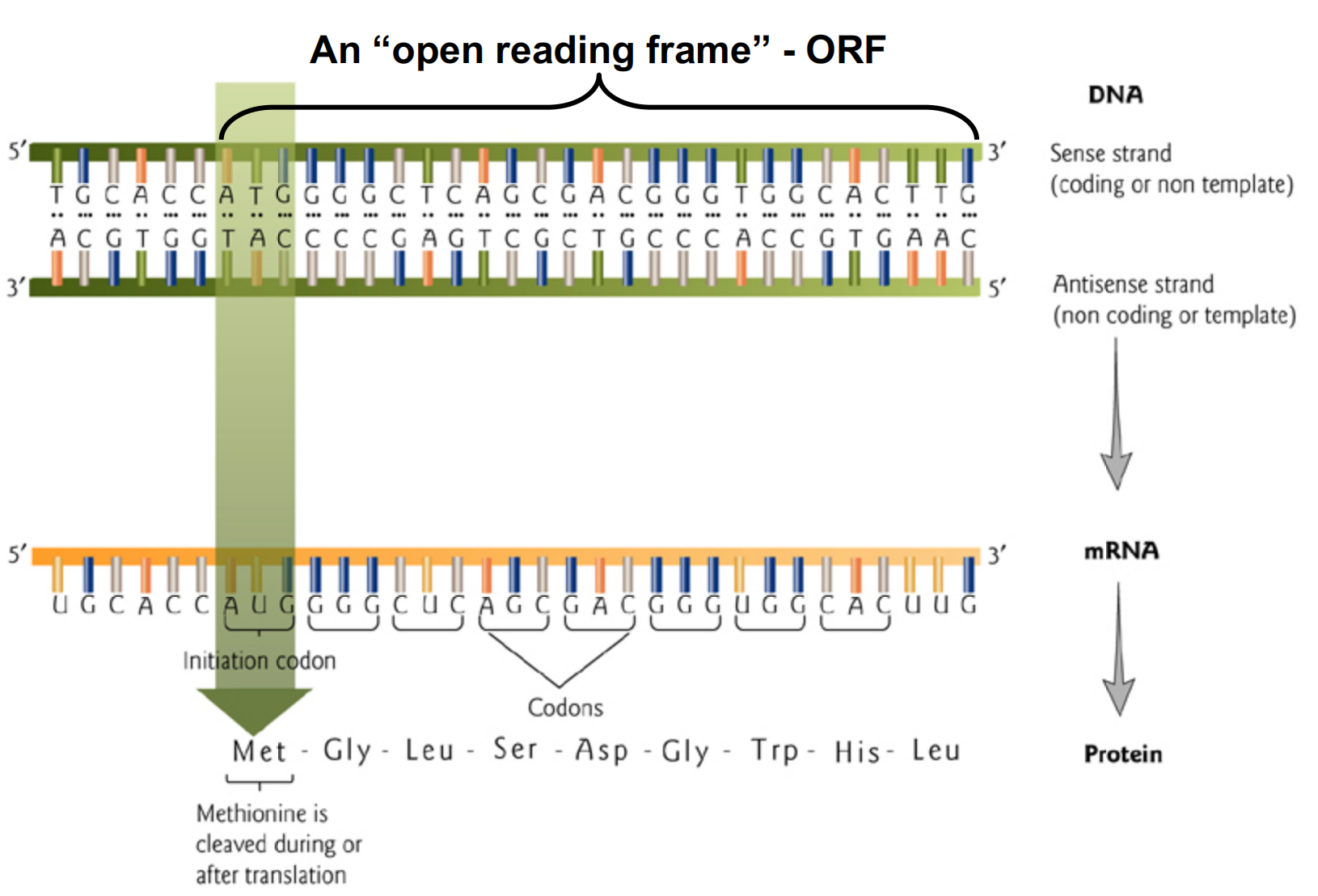
1. Open Reading Frame
In molecular genetics, an open reading frame (ORF) is the part of a reading frame that has the potential to code for a protein or peptide. An ORF is a continuous stretch of codons that do not contain a stop codon (usually UAA, UAG or UGA).
开放阅读框(英语:Open reading frame;缩写:ORF;其他译名:开放阅读框架、开放读架等)是指在给定的阅读框架中,不包含终止密码子的一串串行。这段串行是生物个体的基因组中,可能作为蛋白质编码串行的部分。基因中的ORF包含并位于开始编码与终止编码之间。
2. The Three-bases Code
The Crick-Brenner experiments demonstrated that a set of three nucleotides encodes one amino acid:
Proflavin induced single base changes caused “frameshift” mutations.
Frameshift mutations cause protein “garble” downstream of first base insertion or deletion. After 2nd insertion/deletion, still garbled…but third insertion restores the correct protein.
Thus, three nucleotides = one amino acid
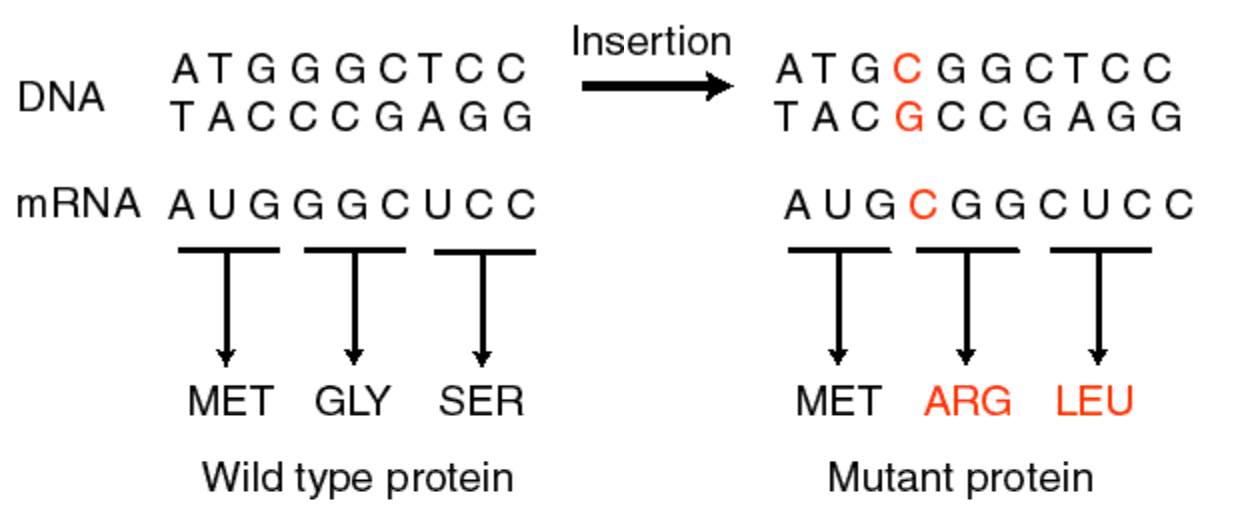
20 amino acids plus stop codons encoded by three-nucleotide sets. The genetic code is “universal” which is to say that it is used in all organisms (with a few exceptions).
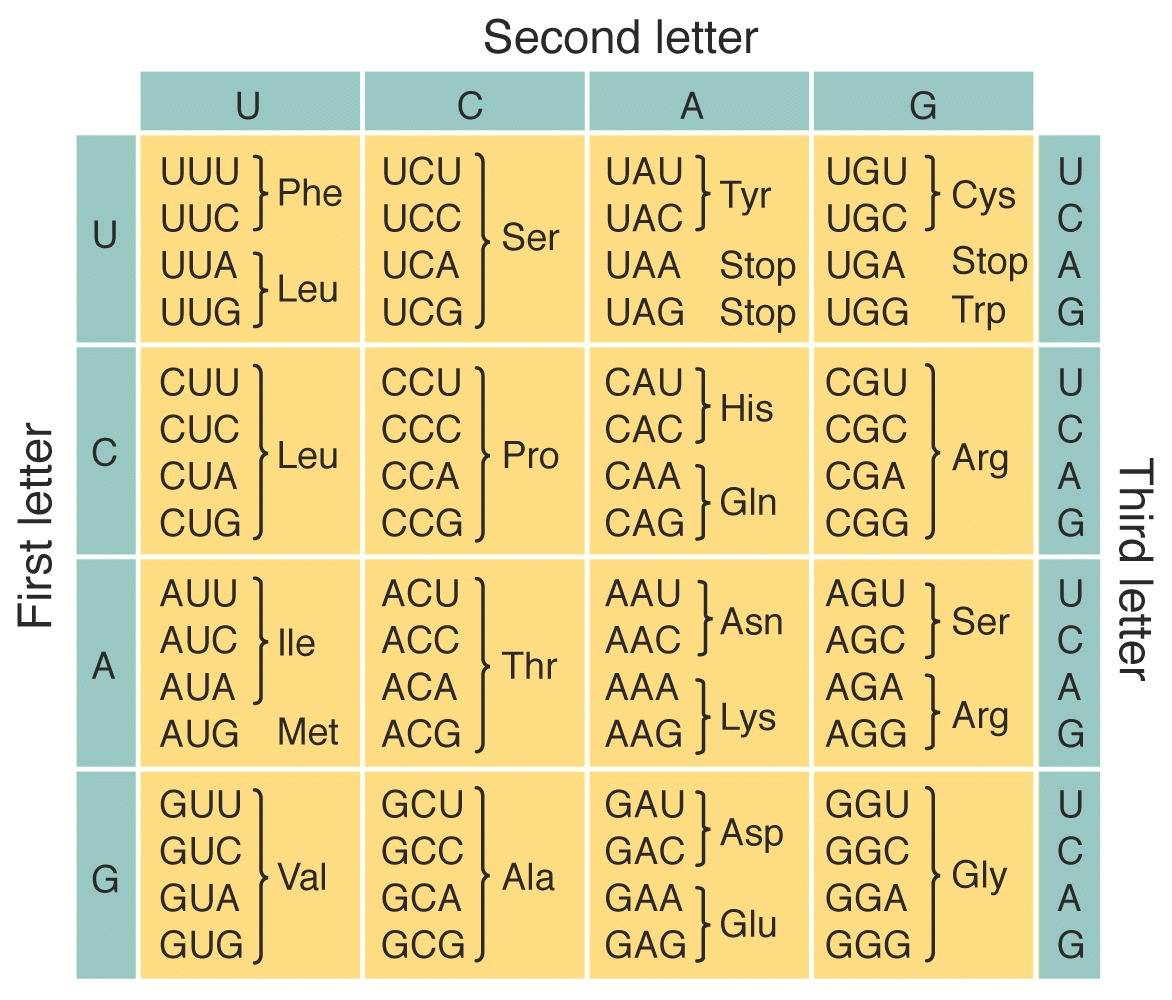
The code is also non-overlapping – each codon has only one use in an organism.
Experiments to show this: Nirenberg/Khorana/Holley experiments (1968 Nobel prize)
Nirenberg & Matthaei: in 1961 used poly-U RNA in cell free extract to make phenylalanine
Khorana et al. used UCUCUCU repeats to produce serine (UCU) and leucine (CUC)
Holley et al. in 1964 isolated tRNA and showed how a codon read and converted to an amino acid
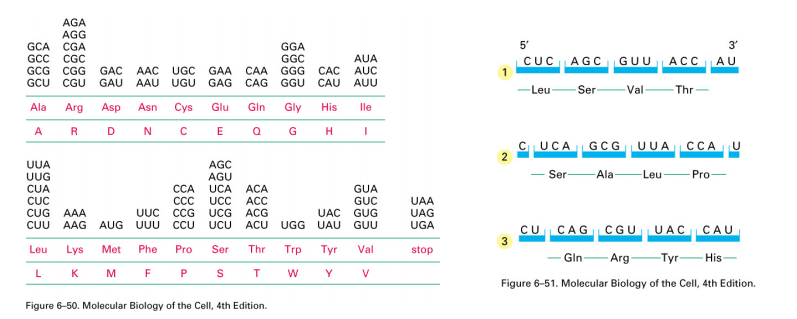
DNA is turned into RNA (‘transcription”), and then the RNA is read during the process of translation (in the 5’ to 3’ direction). Translation uses sets of three bases and converts the information into one amino acid. Thus, 64 possible triplets form 20 amino acids.
The code is “degenerate” or “redundant”.
该code是“简并”或“冗余”的。
Amino Acids
20 amino acids encoded
Grouped by the physiochemical properties of the side chains (“R group”)
Two of these, selenocysteine and pyrrolysine are infrequently utilized, and are NOT found in plants (substituted for two stop codons in genetic code)
1. Acid-base properties of amino acid
The charge of an amino acid can vary depending on the pH
- this is the basis for amino acid identification and protein separation.
2. Peptide bond formation
Condensation reaction to produce polypeptide with a common backbone and variable side chains.
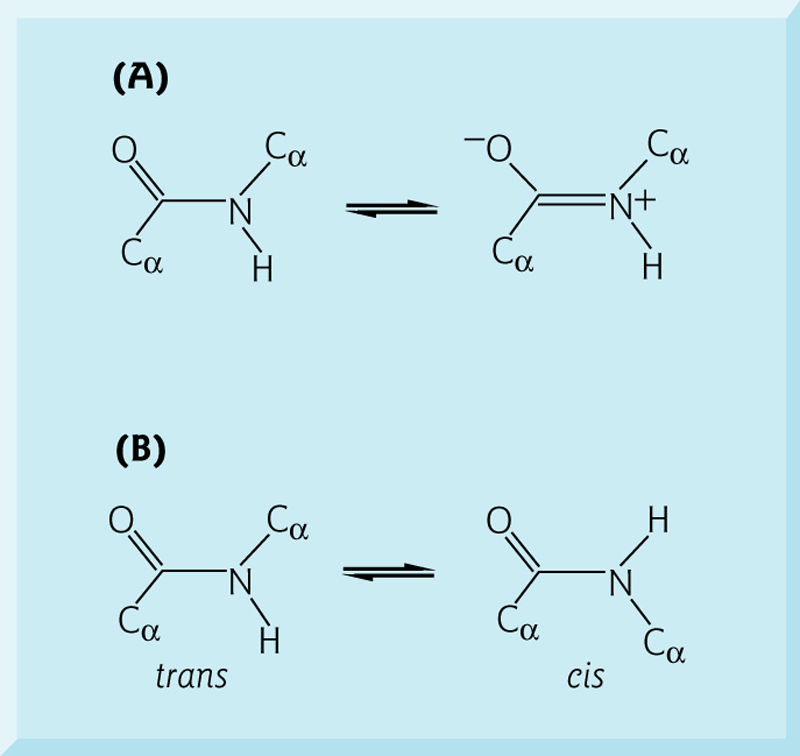
The peptide bond acts as a partial double bond, imparting rigidity.(肽键起部分双键的作用)
Protein
Proteins are also polymers, but have a higher level of complexity than DNA 一 there is no universal structure, although each species of protein has a unique 3-D structure.
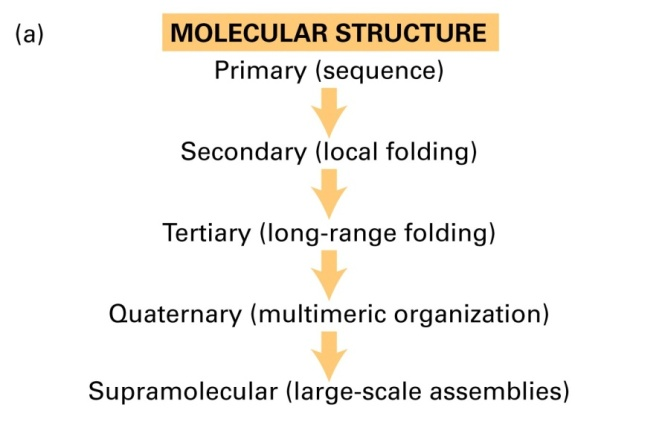
1. Disulfide Bond
Disulfide bonds lock in the secondary structure of a protein via covalent linkages.
Chymotypsin (胰凝乳蛋白酶) contains five disulfide bonds.
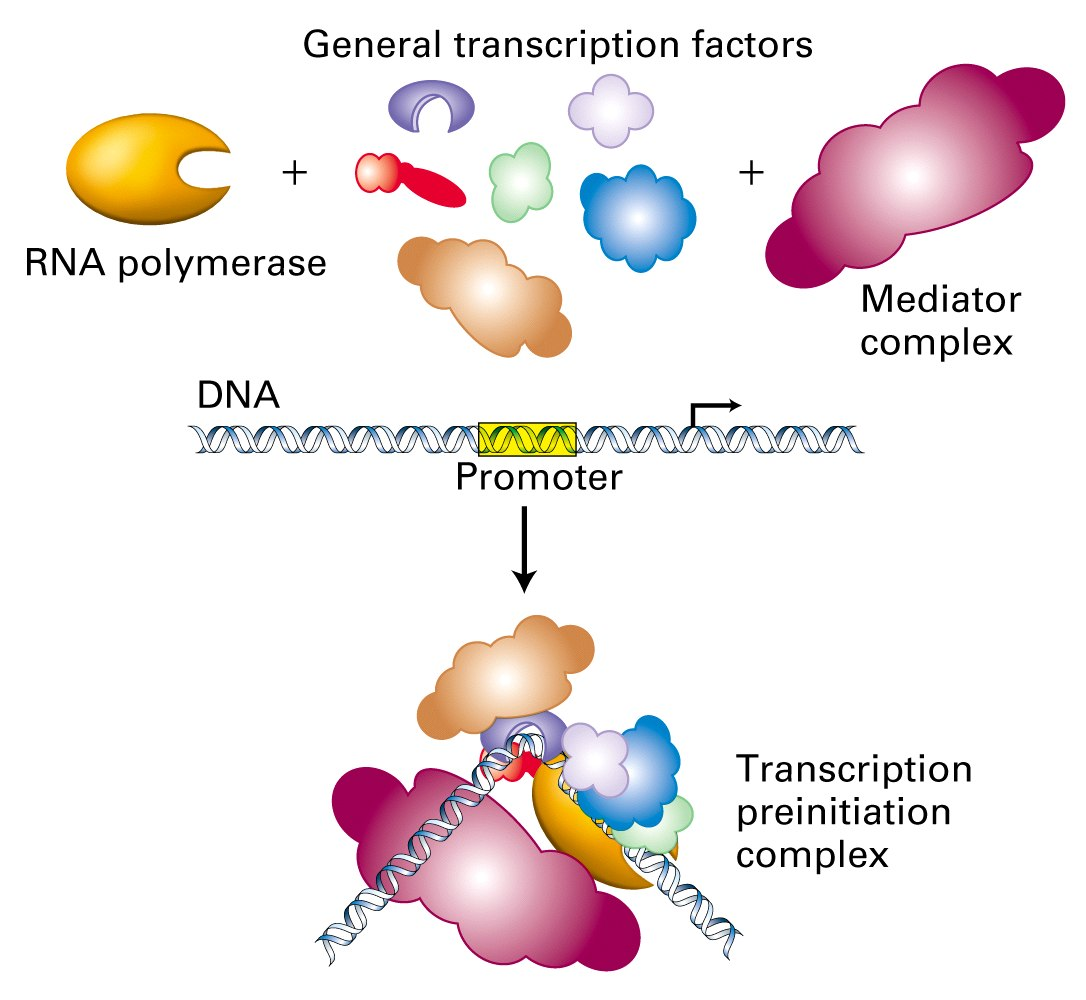
2. Quaternary structure
Quaternary structure defines the number and relative positions of the subunits in a multimeric protein complex. Each protein has a defined role, and the complex may not function without one part.
Proteins are highly adaptable molecules and vary greatly in size, shape and function.
20% of cellular weight is protein.
3. Enzymes
Enzymes catalyze chemical reactions, and can increase the reaction rate by $10^8$ to $10^{10}$ or even higher.
4. Regulation of Protein Activity
Proteins may be produced – but stay in an inactive form
Allosteric Regulation
The allosteric regulation is involved in the regulation of protein activity.
Phosphorylation/Dephosphorylation
A common cellular mechanism for the regulation of protein activity. Phosphorylation can activate or deactivate proteins.
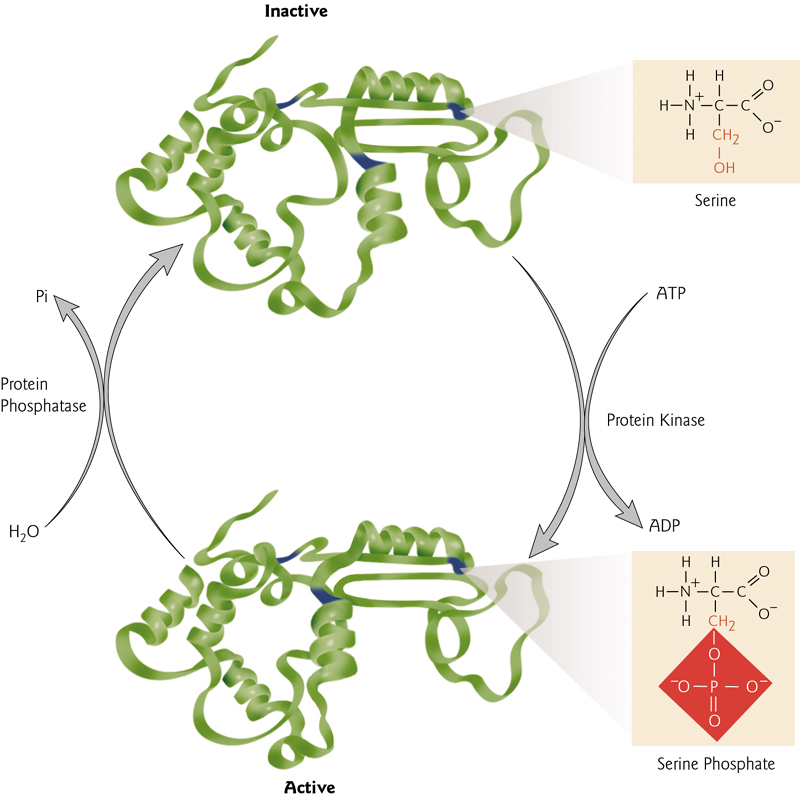
Cyclin-dependent kinases (CDKs,周期蛋白依赖性激酶)
是一个蛋白质激酶家族,因其在细胞周期中的调控作用而首次被发现,该蛋白家族也涉及转录调控、mRNA加工和神经细胞的分化。这类酶存在于已知所有真核生物中。
The activation of CDKs is caused by cyclin bonding and phosphorylation.
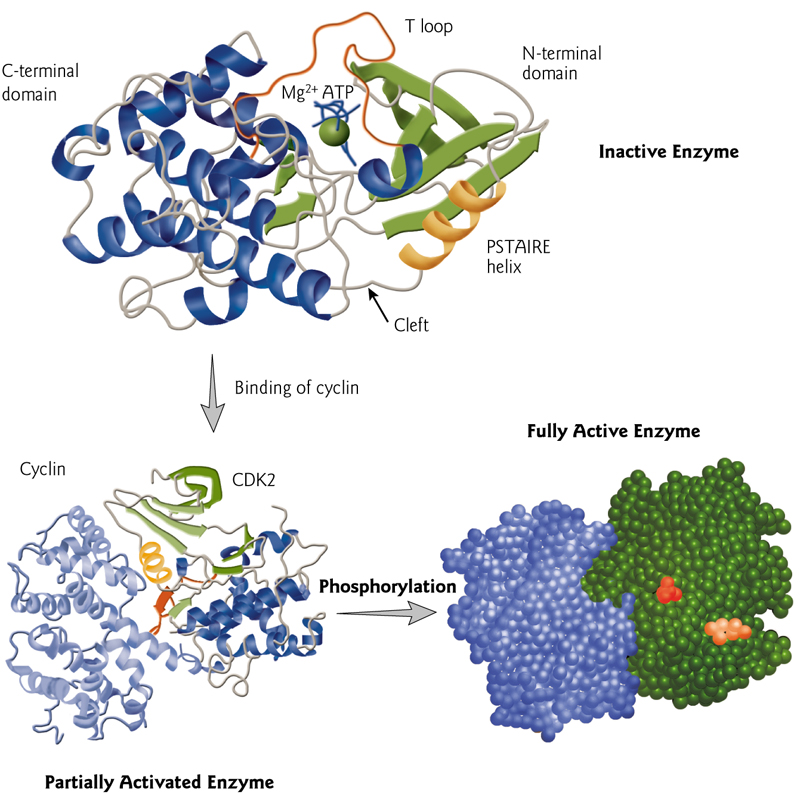
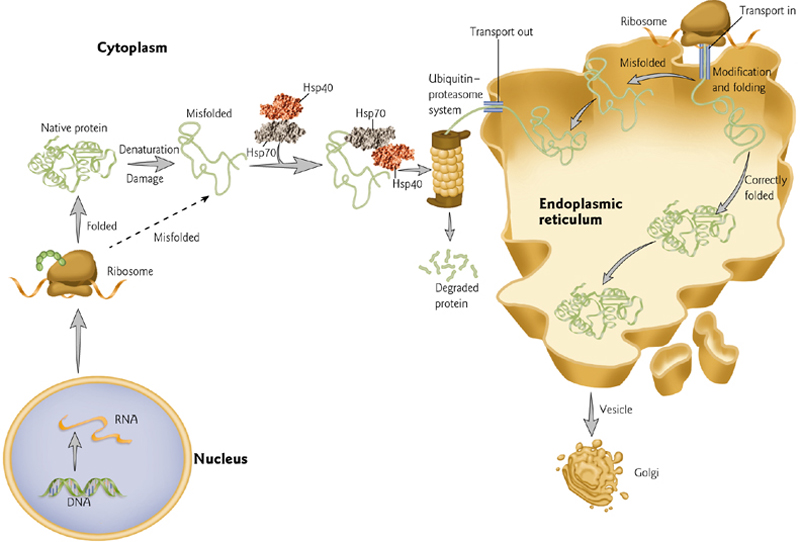
5. Regulation of Protein Folding
Molecular chaperones Hsp70 and Hsp40 assist folding of newly synthesized proteins
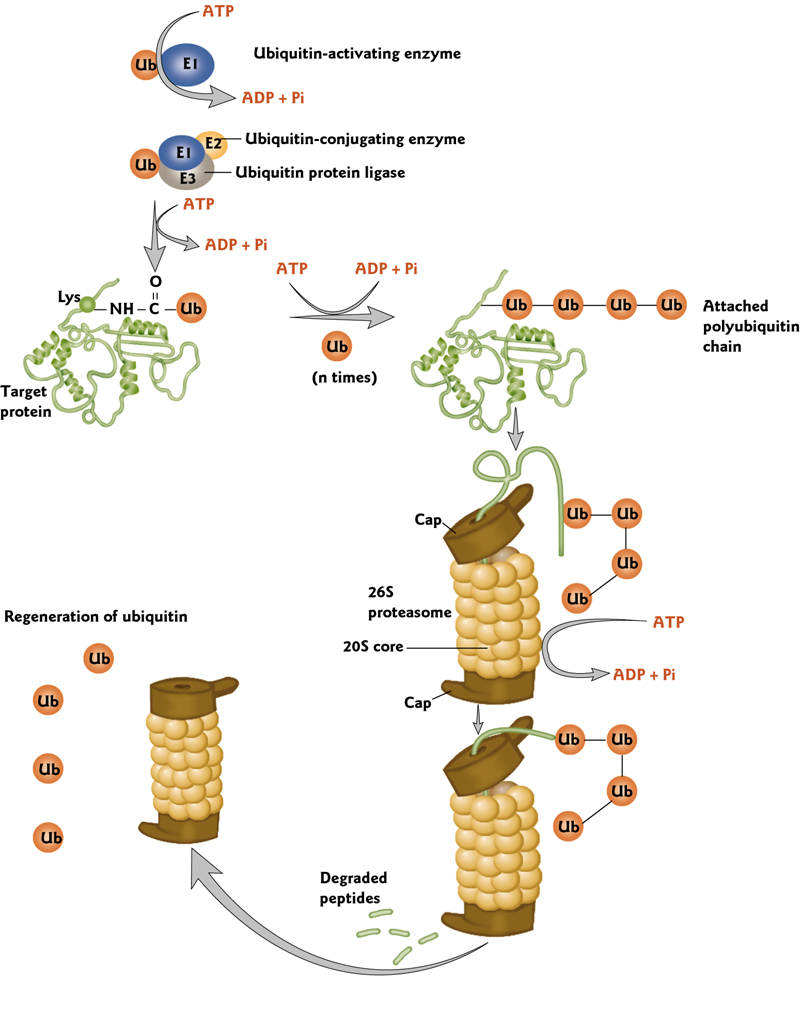
6. Ubiquitin(泛素)-mediated Protein Denaturation
Proteins may be “tagged” for selective destruction in proteolytic complexes(蛋白水解复合物) called proteasomes(蛋白酶体)
This tagging occurs by covalent attachment of ubiquitin, a small, compact and highly conserved protein.
四、Work with Proteins
The key to working with proteins are techniques that allow the biochemist to:
- Separate distinct proteins
- Differentiate or identify distinct proteins after separation
- Concentrate and/or purify distinct proteins while retaining the function (e.g. the correct folding cannot be lost)
- Measure activity.
Knowing the structure of a protein can tell you a great deal about the function and how that protein interacts with other molecules (ligands or interacting proteins).
If the proteins are in complexes with other proteins, it is important to know:
- What the other interacting proteins are
- The number of interacting proteins
- The relative ratios of each protein in the complex (the “stoichiometry”).
- How they are assembled relative to one another.
And if the proteins are interacting with non-protein molecules, it’s important to know what they are (e.g. small molecules, DNA, RNA, etc).
Protein Separation
1. Centrifugation
Centrifugation is a very important method that allows you to separates molecules of different sizes.
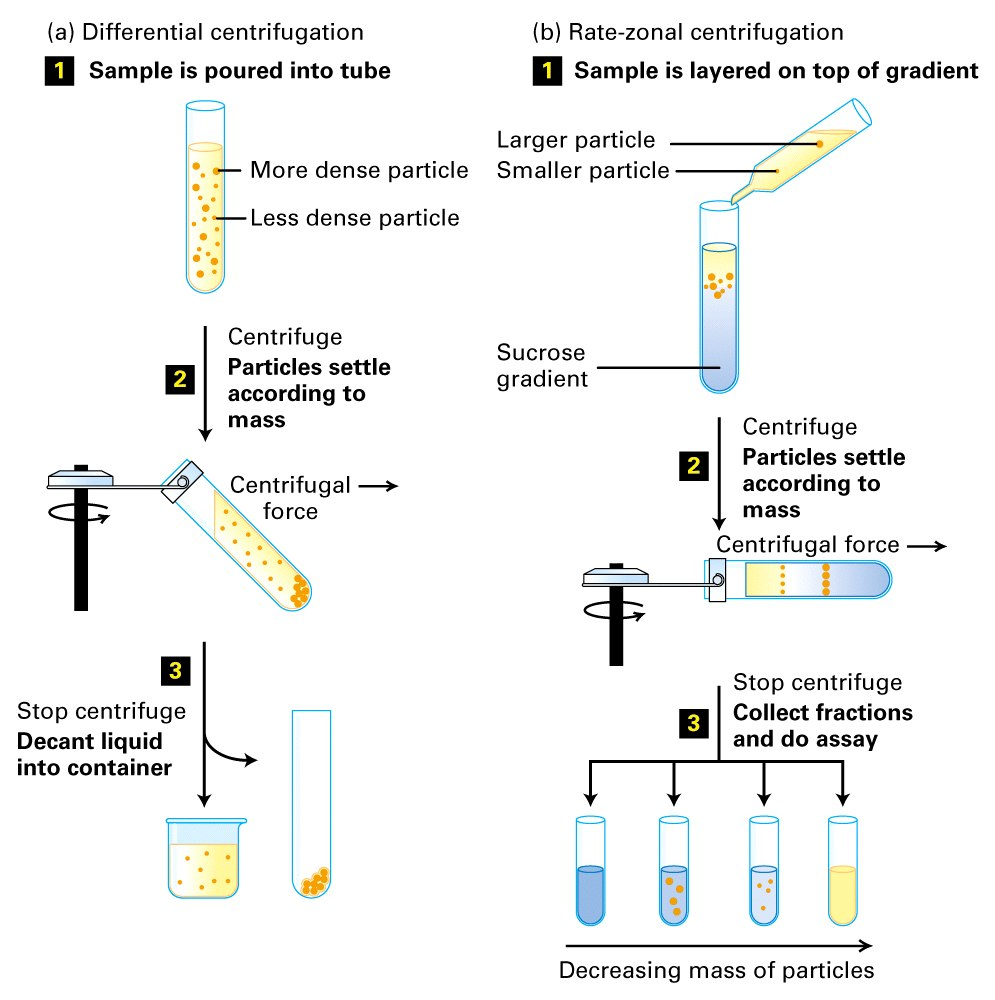
Separation of Cellular Components
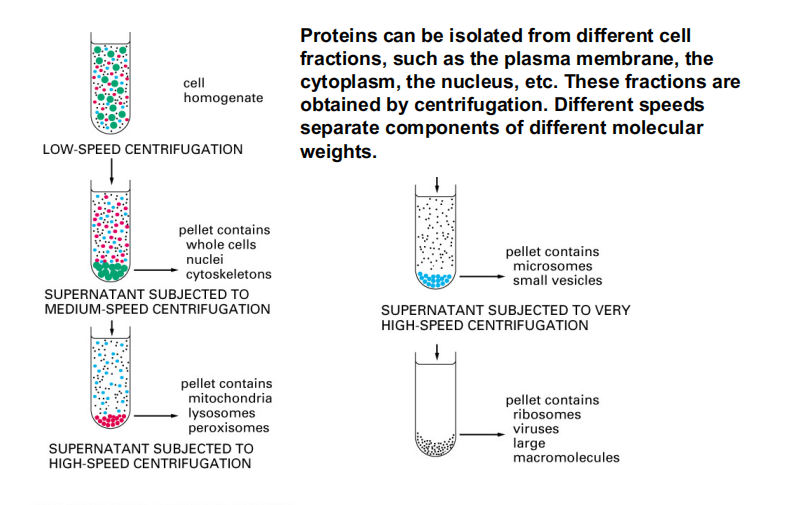
Proteins can be isolated from different cell fractions, such as the plasma membrane, the cytoplasm, the nucleus, etc. These fractions are obtained by centrifugation. Different speeds separate components of different molecular weights.
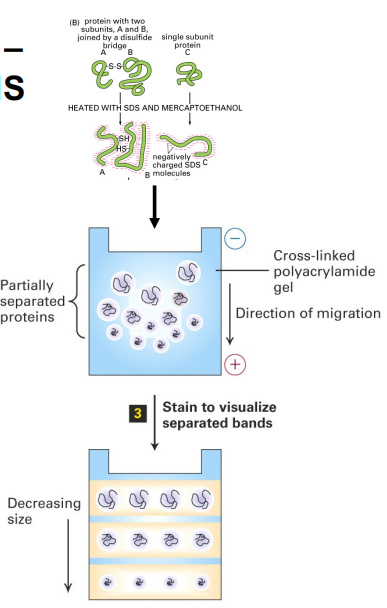
2. Gel Electrophoresis
Protein are separated on poly-acrylamide gels(聚丙烯酰胺凝胶).
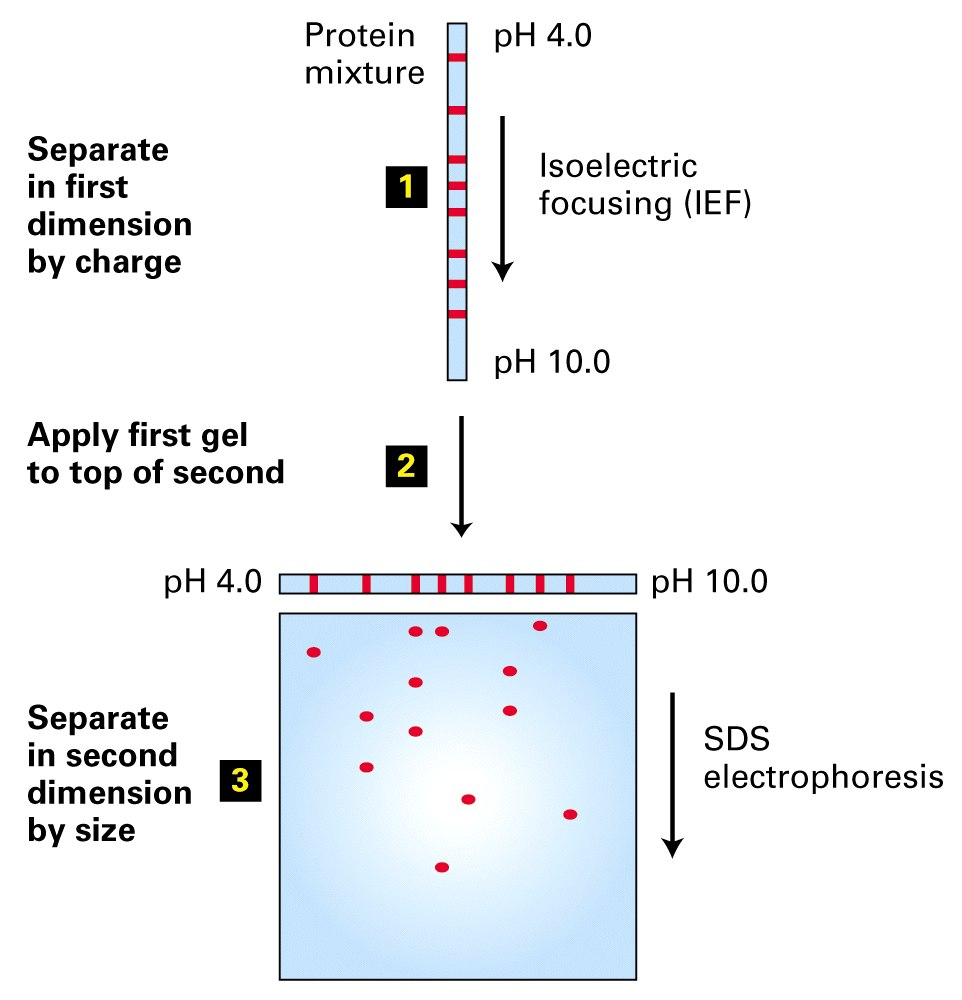
2-D Gel Electrophoresis
Because of the different amino acid side chains that are exposed on the surface of the protein, proteins have different charges.
These charges can be used to separate proteins by isoelectric focusing in a pH gradient.
The combination of isoelectric focusing and SDS gel electrophoresis can separate proteins in two dimensions, hence the “2-D gel”.
Separation of Cellular Components
Proteins can be isolated from different cell fractions, such as the plasma membrane, the cytoplasm, the nucleus, etc. These fractions are obtained by centrifugation.
Different speeds separate components of different molecular weights.
Translation
Electron micrographs of the small and large ribosomal subunits.
These are ribonucleoprotein complexes, a polypeptide polymerase essentially
Ribosomes are comprised of the two subunits, which each have a ribosomal RNA and numerous protein components. Ribosomes convert the genetic code of the mRNA into proteins, catalyzing the formation of polypeptide bonds.
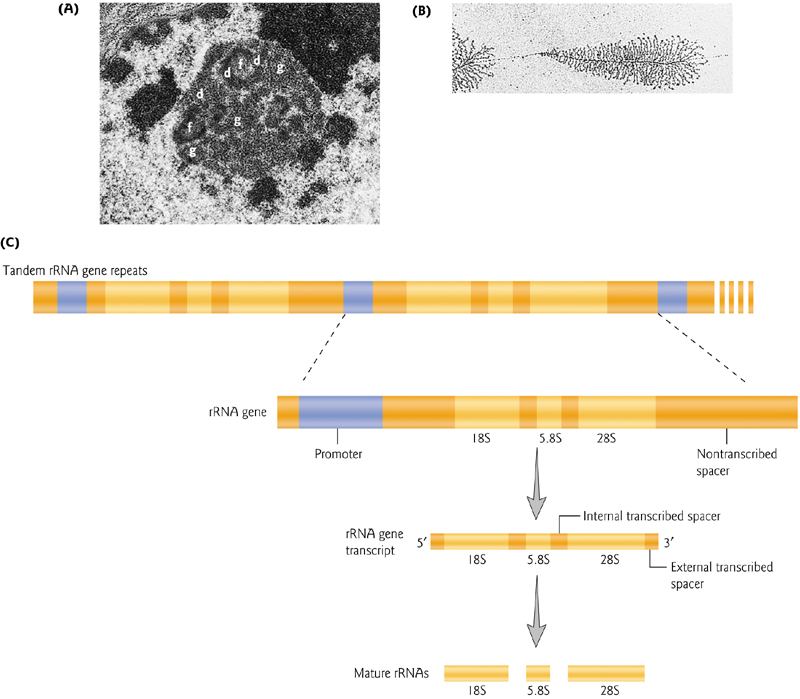
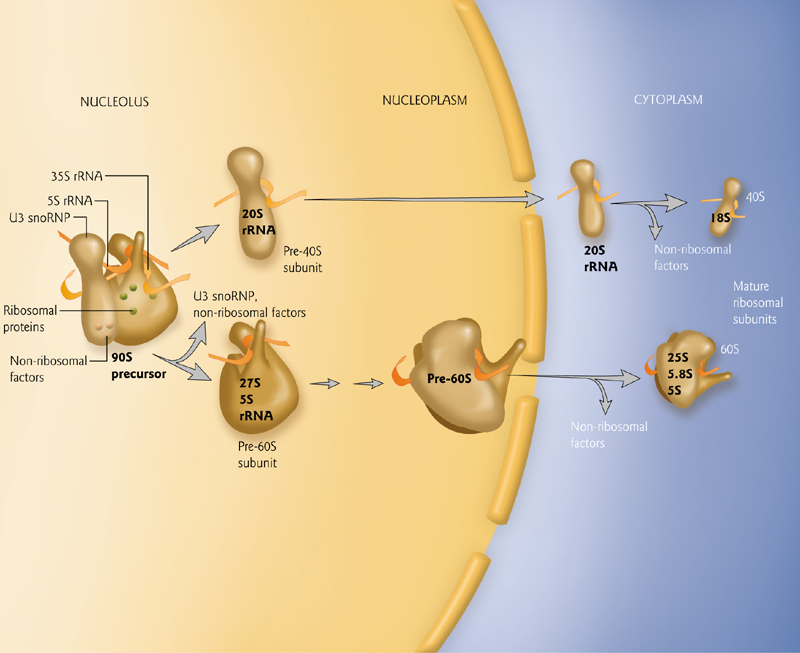
1. The Nucleolus is the site of ribosomal production from high copy gene clusters
Tandem sets of thousands of rRNA genes are required for the production of the many ribosomes used in protein production.
This ribosome production takes place in the nucleoleus, a visible cellular compartment.
rRNAs are transcribed by RNA pol I, with the exception of the 5S rRNA, transcribed by RNA pol III.
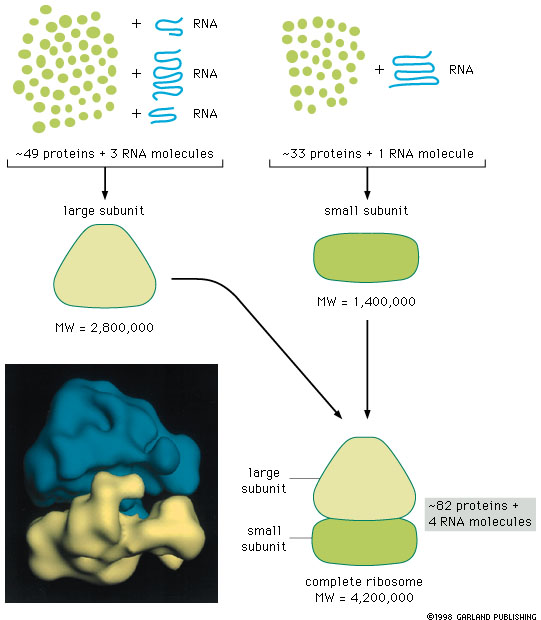
2. Ribosome Composition
Eukaryotic example: Two subunits (40S and 60S), with variation in sizes across organisms. The 40S contains an 18S rRNA, while the 60S contains a 5S, 5.8S and 28S rRNA.
The 5S rRNA is synthesized in the nucleus, and the other rRNAs in the nucleolus.
3. Ribosome Production
Subunits are produced in the nucleolus, matured in the nucleus, and exported to the cytoplasm where they are used in protein production
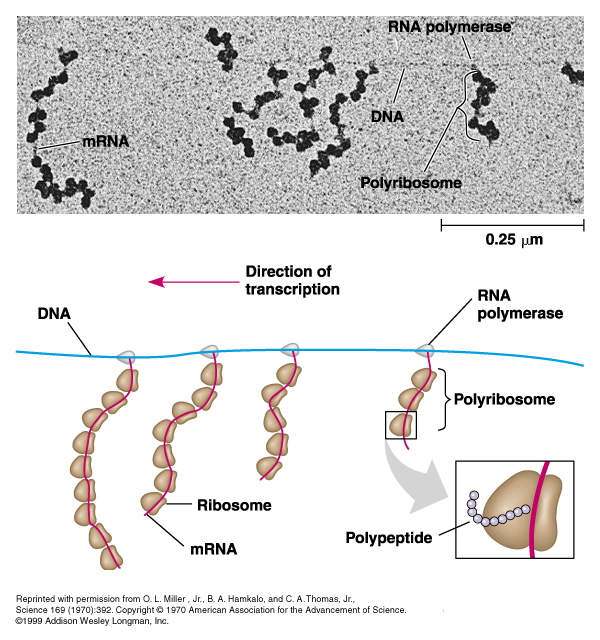
4. Polyribosomes
Prokaryotes can simultaneously transcribe and translate genes;
Eukaryotes can simultaneously produce multiple proteins with multiple ribosomes (polysomes), but transcription is separate because the mRNA is processed.
tRNA
tRNAs provide the connection from a codon of three bases to a single amino acid
There is “degeneracy” in the anticodon 3rd position, so 61 different tRNAs are not necessary.
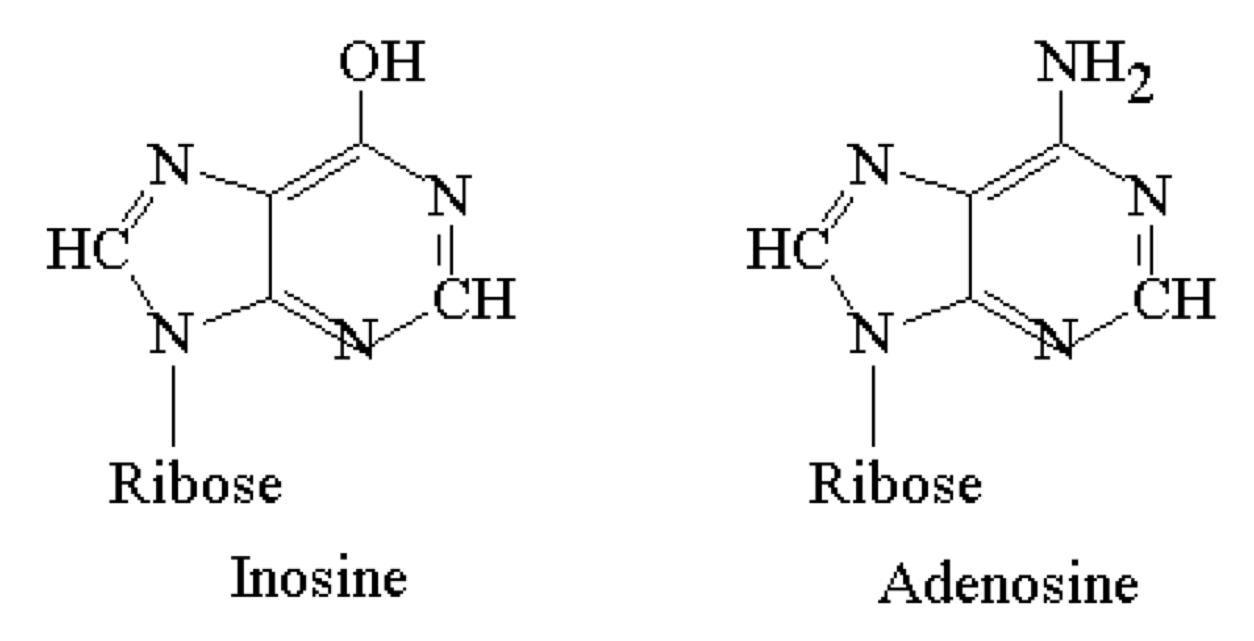
tRNA anticodons may have inosine in this 3rd position to facilitate degeneracy
- Inosine can pair with U, C, or A and is thus “wobbly”
- Looks like adenosine
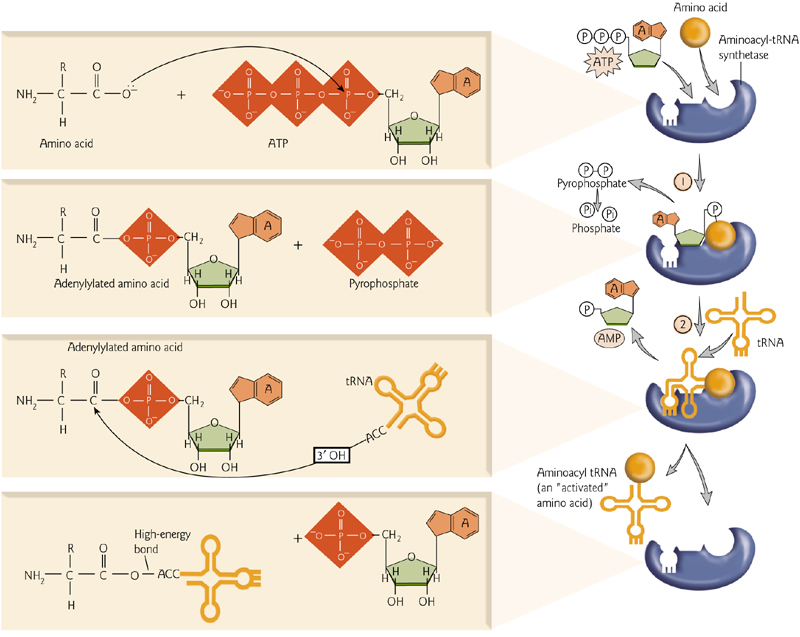
1. ‘Charge’ tRNA
tRNAs are “charged” when the correct amino acid is added to the RNA component.
This requires ATP and an important enzyme, an aminoacyl-tRNA synthetase.
2. Working tRNA
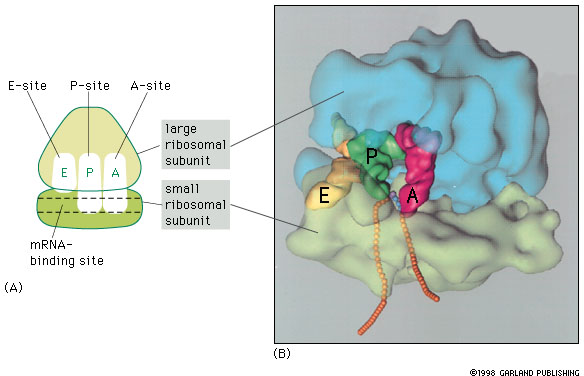
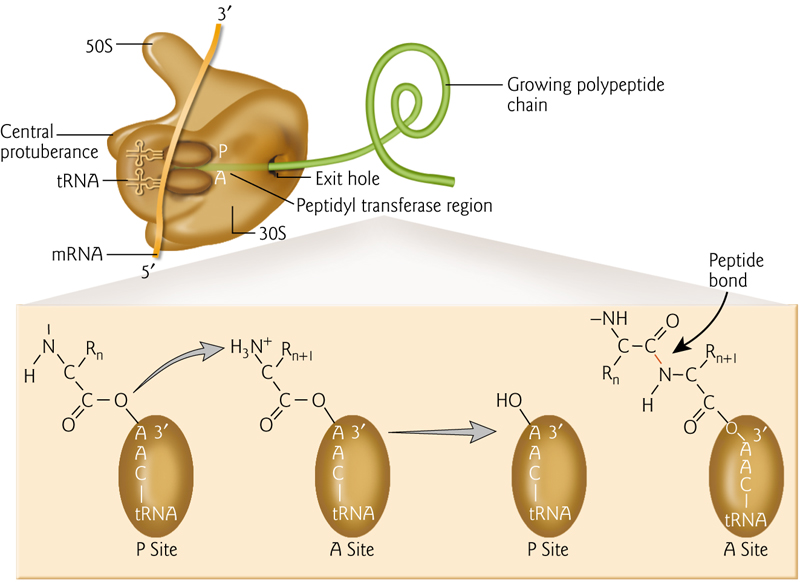
RNA-binding sites in the ribosome include mRNA and 3 binding sites for tRNA (A-, P-, and E-sites, short for aminoacyl-tRNA, peptidyl-tRNA, and exit).
All 3 tRNA sites are shown occupied below, but during protein synthesis only two of these sites contain tRNA molecules at any one time
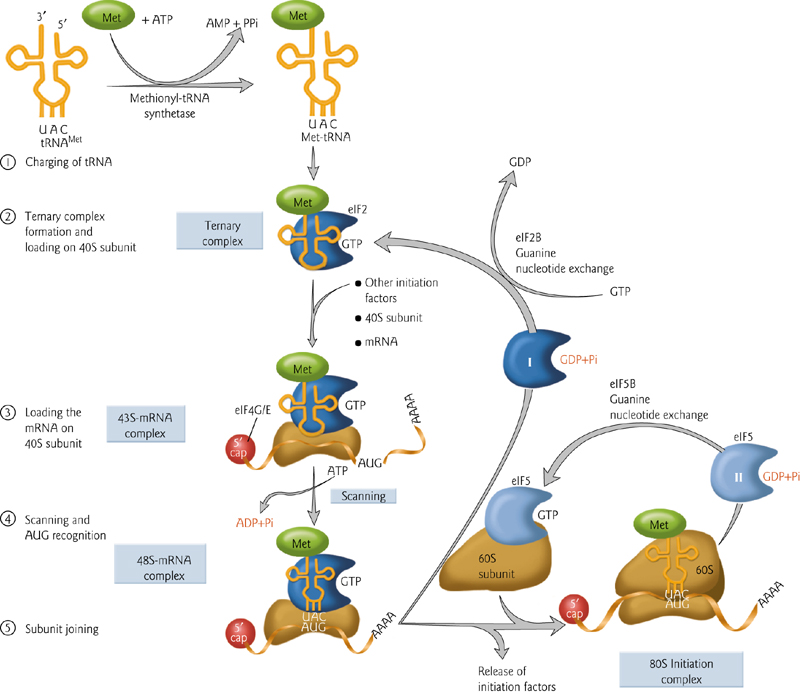
3. Translation Initiation – Start Codon
The first amino acid is different – there is nothing to attach it to (no growing chain of amino acids). Therefore this is handled different than the rest. Initiation factors are required.
The secondary structure of the 5’ cap is unwound to facilitate translation. The proteins that attach to this cap promote translation.
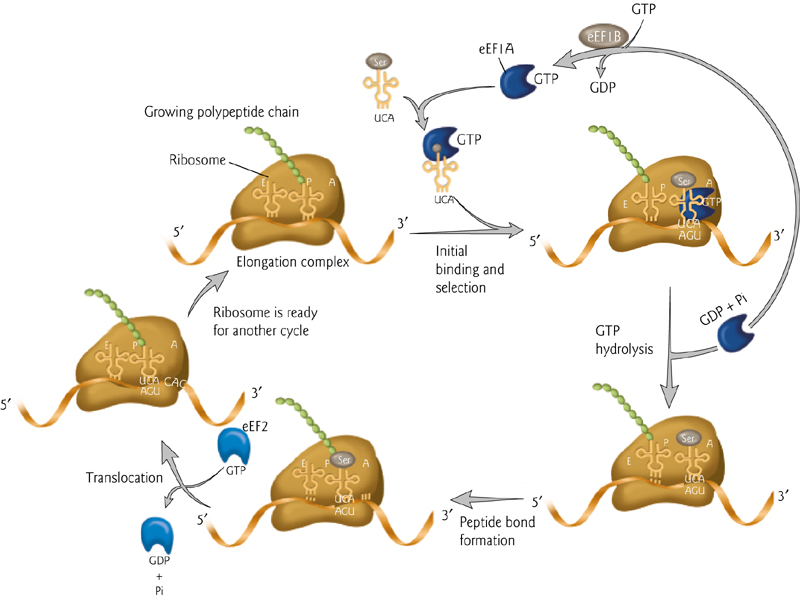
Translating an mRNA Molecule
3-step cycle repeated during protein synthesis.
An aminoacyl-tRNA molecule binds to the A-site on the ribosome in step 1
A new peptide bond is formed in step 2,
The small subunit moves a distance of three nucleotides along the mRNA chain in step 3, ejecting the spent tRNA molecule and “resetting” the ribosome
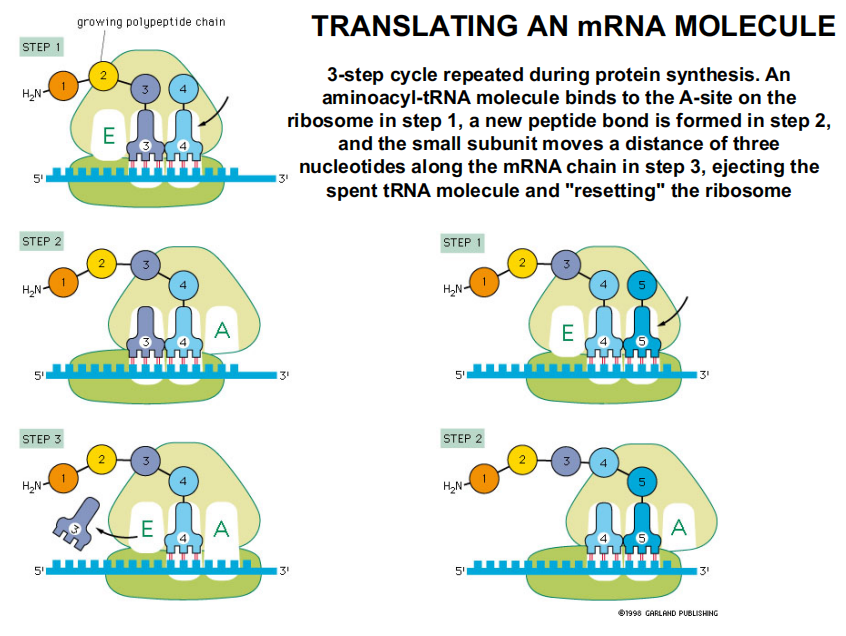
In the Allison textbook version:
An aminoacyl-tRNA (氨酰tRNA) molecule binds to the A-site on the ribosome, forms a new peptide bond, then the small subunit moves three nucleotides along the mRNA, “resetting” the ribosome.
4. Formation of The Peptide Bond
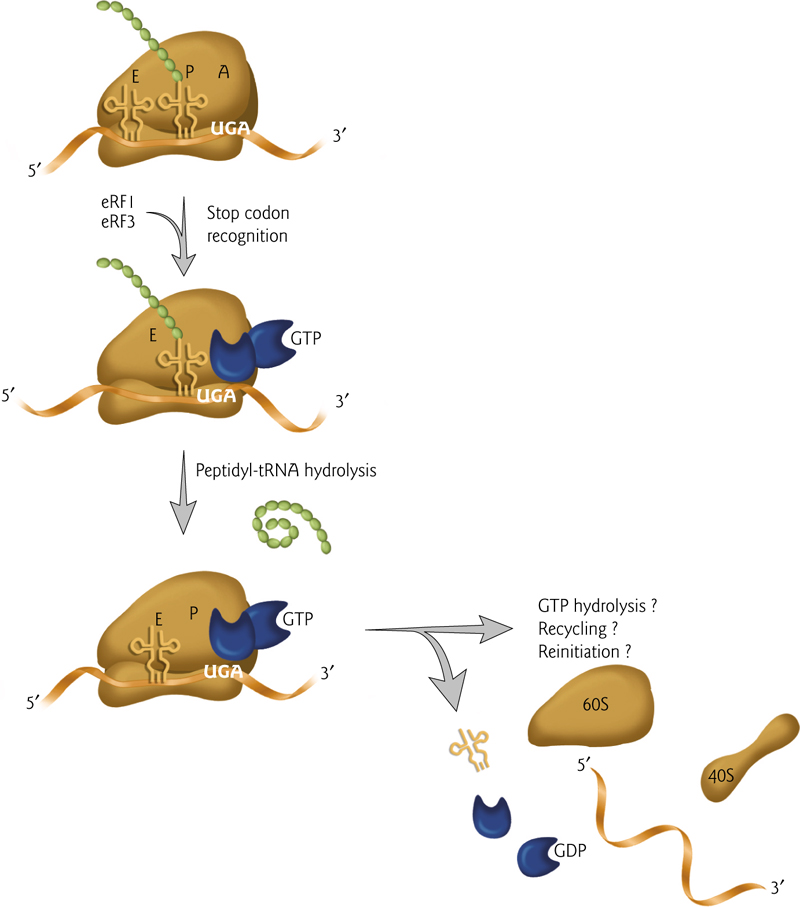
5. Termination of Translation
Translation ends when the stop codon is reached; no additional peptide bonds can be formed.
The last step is the hydrolysis of the peptide chain from the tRNA.
Ribosomal parts can then be recycled or can restart the process of translation.