L9 DNA&RNA Modifying Enzymes
一、Polynucleotide Kinase & Alkaline Phosphatase
T4 polynucleotide Kinase
Origin: T4 phage
Feature:
- Catalyzes the transfer of the gamma-phosphate from ATP to the 5’-OH group of singleand double-stranded DNAs and RNAs.
- The enzyme is also a 3’-phosphatase under appropriate condition
Usage:
- 5’ phosphorylation of DNA/RNA for subsequent ligation
- End labeling DNA or RNA for probes and DNA sequencing
- Phosphorylation of PCR primers
- Removal of 3’ phosphoryl groups

Alkaline Phosphatase
Origin: Calf or Shrimp
Feature:
- Alkaline Phosphatase catalyzes the release of 5’and 3’-phosphate groups from DNA, RNA, and nucleotides.
- This enzyme also removes phosphate groups from proteins
- CIAP (Calf intestinal AP) & SAP (Shrimp AP).
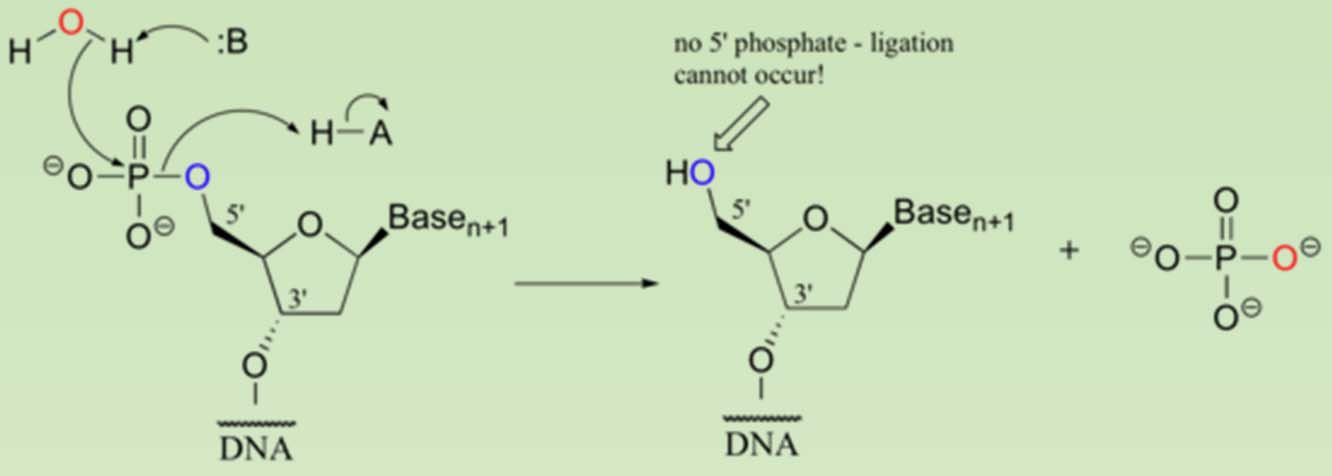
- Phosphatase needs to be inactivated before the DNA is used for ligation.
Usage:
- removes 3´ and 5´ phosphate group from DNA, RNA and dNTP
- To prevent self-ligation
二、Terminal Deoxynucleotidyl Transferase & Poly(A) Polymerase
Terminal Deoxynucleotidyl Transferase
Origin: Calf thymus
Feature:
- TdT is a template-independent DNA polymerase that catalyzes the repetitive addition of deoxyribonucleotides to the 3’-OH of oligodeoxyribonucleotides and single-stranded and double-stranded DNA
**Usage: **
- To create single-stranded overhang of dsDNA – for gene cloning
- DNA 3’end labling.
- Creats a sticky end

Poly(A) Polymerase
Origin: E.coli or Yeast
Feature:
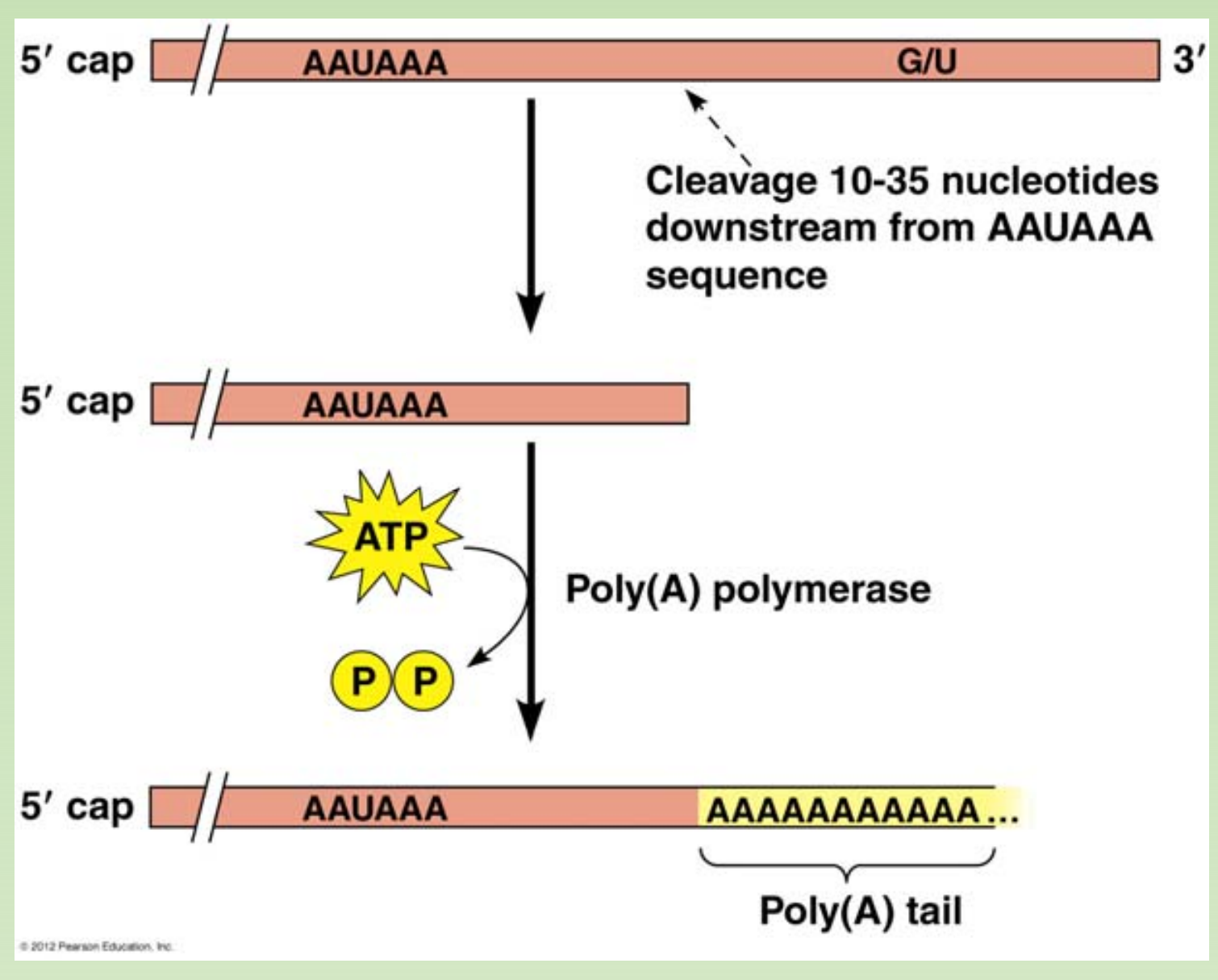
- catalyzes the addition of adenosine to the 3’ end (-OH) of RNA in a sequence-independent fashion
**Usage: **
- Labeling RNA 3’ end; facilitate reverse transcription, cloning & sequencing.
三、Nucleases – DNase I, Exonuclease I, RNase A, RNase I, RNase, H, MNase
Nucleases – DNase I
Origin: E.coli or Yeast
Feature:
- A nonspecific endonuclease that degrades dsDNA, ssDNA and DNA in RNA:DNA hybrid.
- It functions by hydrolyzing phosphodiester linkages, producing mono and oligonucleotides with a 5’-phosphate and a 3’-hydroxyl group
Usage:
- Preparation of DNA-free RNA.
- DNA labeling by nick-translation in conjunction with DNA Polymerase I
- Studies of DNA-protein interactions by DNase I footprinting
DNase I foot-printing to study DNA-protein interaction:

Exonuclease I (E. coli)
Origin: E.coli
Feature:
- 3’ to 5’ exonuclease of ssDNA
- Catalyzes the removal of nucleotides from linear single stranded DNA in the 3’ to 5’ direction

Usage:
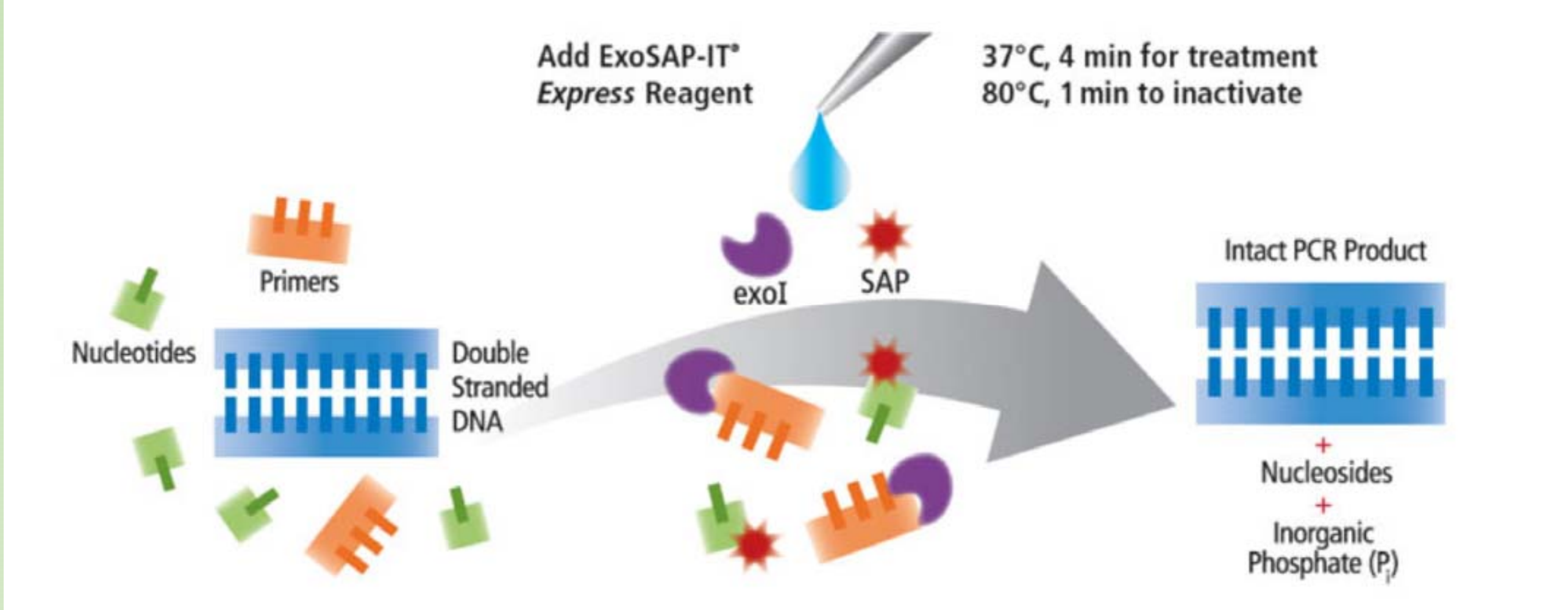
- Remove excessive primers from PCR product
- Exo-SAP treatment
- The exonuclease I removes leftover primers, while the Shrimp Alkaline Phosphatase removes any remaining dNTPs
- Exo-SAP treatment
RNase A
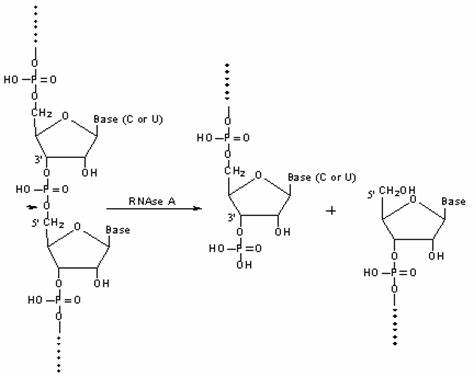
Degrades single-stranded RNA at C and U residues.
Cleaves the phosphodiester bond between the 5’-ribose of a nucleotide and the phosphate group attached to the 3’-ribose of an adjacent pyrimidine nucleotide (C or U).
Leave a 3’ end phosphate group after clevage.
Resistant to heat, easy to refold – the enzyme that we should worry about in RNA extraction.
RNase I
RNase I degrades single-stranded RNA to nucleoside 3´ monophosphates via 2´,3´ cyclic monophosphate intermediates by cleaving between all dinucleotide pairs
The enzyme is completely inactivated by heating at 70°C for 15 minutes.
A more controllable choice to digest RNA than RNase A
RNase H
specifically degrades the RNA strand of an RNA-DNA hybrid to produce 5’ phosphate terminated oligoribonucleotides and single-stranded DNA
Application:
- remove RNA from cDNA

Micrococcal nuclease (MNase)
Exhibits exoand endo-5’-phosphodiesterase activities against DNA and RNA. ds/ss DNA/RNA.
- cut them all
Preference for ATor AU-rich regions.
Strictly dependent on calcium for digestion of RNA and DNA substrates. Precisely inactive the enzyme by adding EGTA
Application: remove RNA from cDNA
- Remove nucleic acids from solution
- Release nucleosome from chromatin
Proteinase K
Cloned from Engyodontium album, is a nonspecific serine protease that is useful for general digestion of proteins.
Optimal activity between 6.5 and 9.5
Active under denaturing conditions, SDS or urea
Active under high temperature, 55-65 degree
Application:
- remove proteins from solution/cell extract (e.g. DNA or RNA extraction)
Summary
- Polynucleotide Kinase (Both a Kinase and phosphatase)
- Alkaline Phosphatase (You need to remove it before ligation)
- Terminal Deoxynucleotidyl Transferase (For add a stretch of A on DNA)
- Poly(A) Polymerase (For add a stretch of A on RNA).
- Nucleases – DNase I, Exonuclease I, RNase A, RNase I, RNase H, MNase (They all cut nucleic acids with different features,think of a scenario that you may use it).