L07 The Small RNA World
一、microRNAs
The Definition
1. Roles and Functions in Cell
- Small RNAs are a pool of 21 to 24 nt RNAs that generally function in gene silencing
- Small RNAs contribute to post-transcriptional gene silencing by affecting mRNA translation or stability
- Small RNAs contribute to transcriptional gene silencing through epigenetic modifications to chromatin
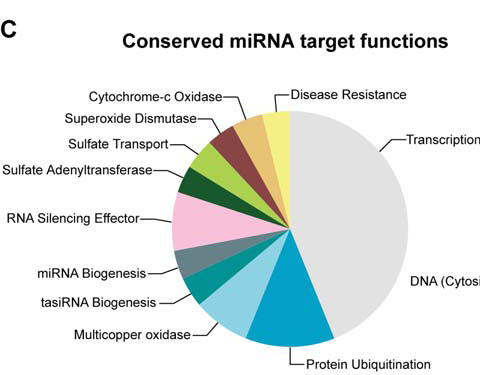
Some miRNAs are highly conserved and important gene regulators
Nearly half of the targets of conserved miRNAs are transcription factors.
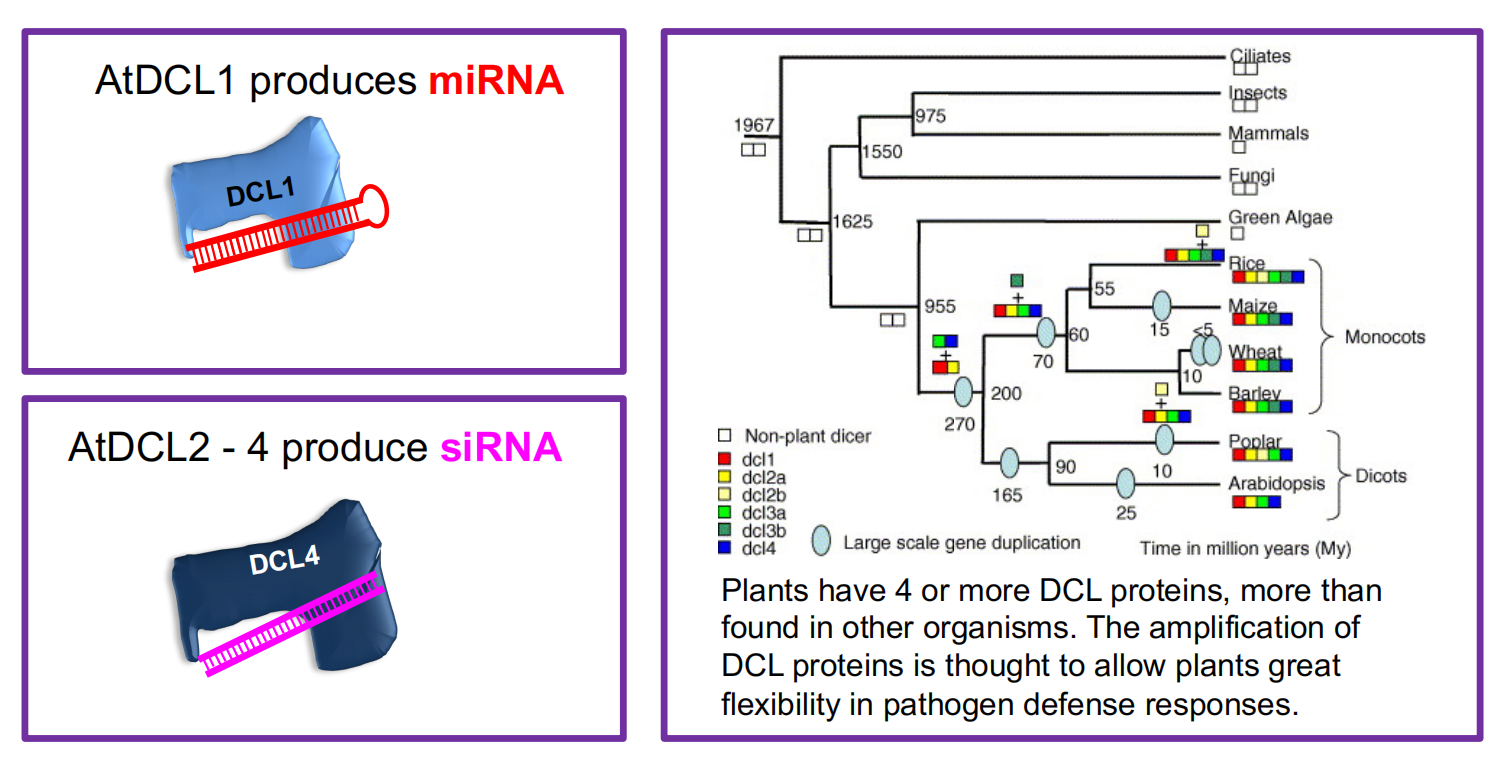
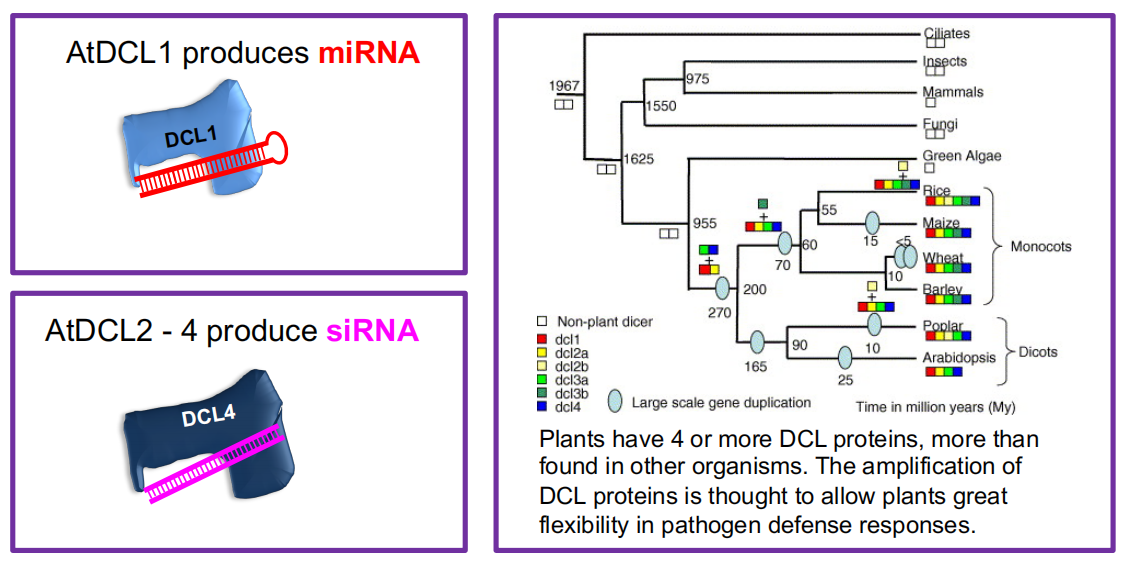
2. Dicers and Argonatures
RNA silencing uses a set of core reactions in which double-stranded RNA (dsRNA) is processed by Dicer or Dicer-like proteins into short RNA duplexes.
These small RNAs subsequently associate with ARGONAUTE proteins to confer silencing.
Dicer and Dicer-like Proteins
In siRNA and miRNA biogenesis, Dicer or Dicer-like (DCL) proteins cleave long dsRNA or foldback (hairpin) RNA into ~ 21 – 25 nt fragments.
(1) miRNAs and siRNAs are processed by related but different DCL proteins
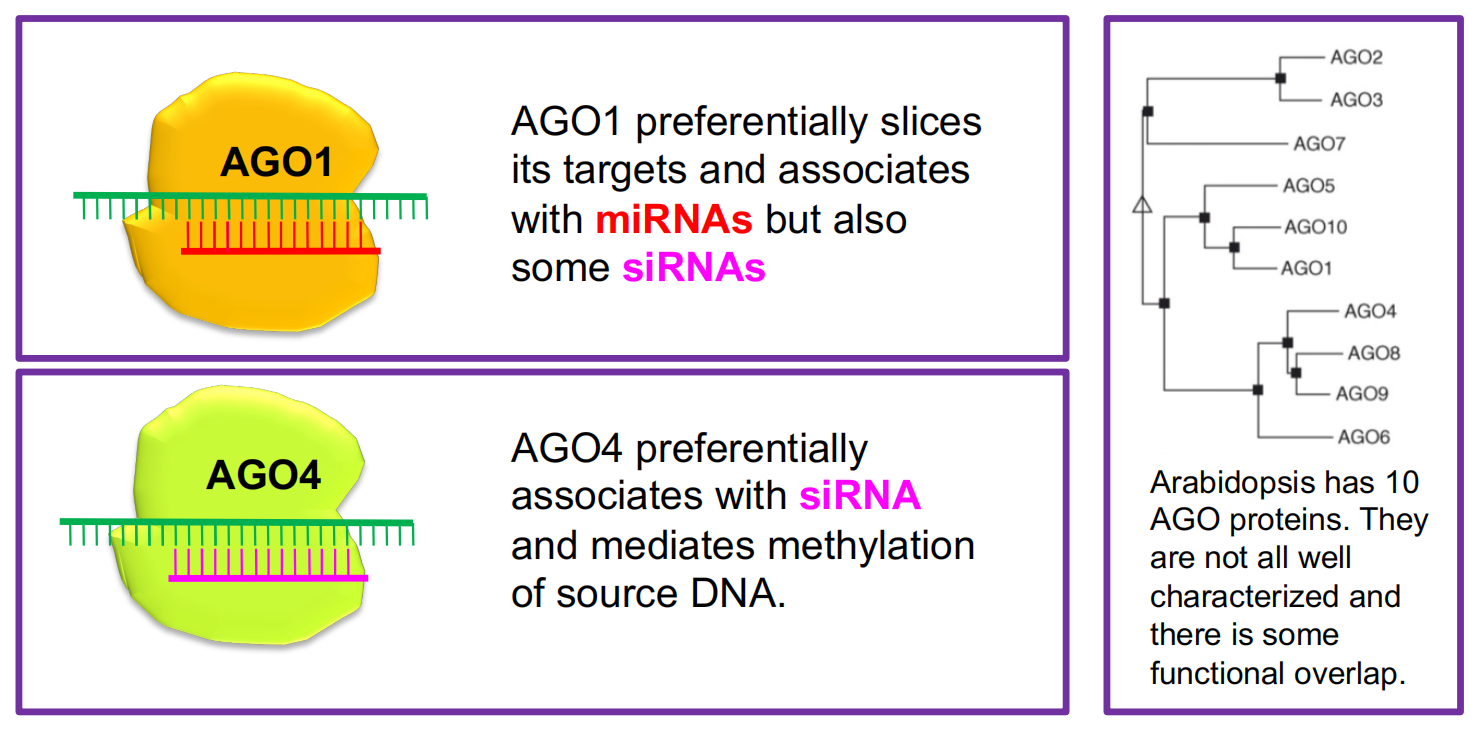
Argonaute proteins
ARGONAUTE proteins bind small RNAs and their targets.
ARGONAUTE proteins are named after the argonaute1 mutant of Arabidopsis; ago1 has thin radial leaves and was named for the octopus Argonauta which it resembles.
(1) miRNAs and siRNAs associate with several AGO proteins
(2) miRNAs and siRNAs are processed by related but different DCL proteins
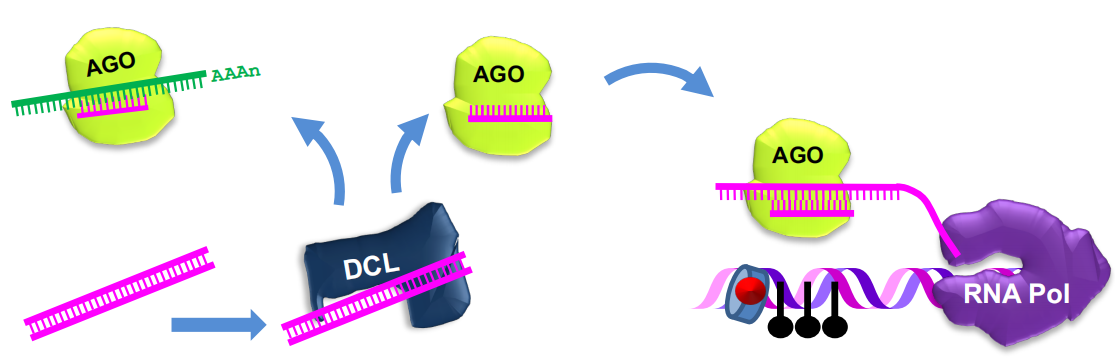
RNA silencing
siRNA -mediated silencing via post-transcriptional and transcriptional gene silencing
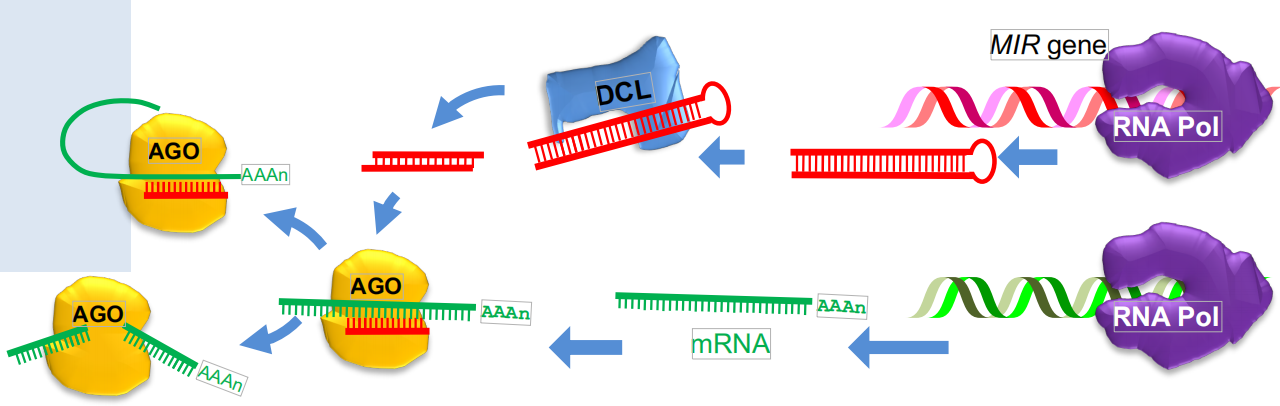
miRNA -mediated slicing of mRNA and translational repression
1. miRNAs
- miRNAs are thought to have evolved from siRNAs, and are produced and processed somewhat similarly
- Plants have a small number of highly conserved miRNAs, and a large number of non-conserved miRNAs
- miRNAs are encoded by specific MIR genes but act on other genes – they are trans-acting regulatory factors
- miRNAs in plants regulate developmental and physiological events
microRNAs slice mRNAs or interfere with their translation
Most siRNAs are produced from transposons and repetitive DNA
- Most of the cellular siRNAs are derived from transposons and other repetitive sequences. In Arabidopsis, as shown above, there is a high density of these repeats in the pericentromeric regions of the chromosome.
Small interfering RNAs (siRNAs) are preferentially derived from pericentromeric region
- The density of small RNA-homologous loci is highest in the centromeric and pericentromeric regions which contain a high density of repeat sequence classes, such as transposons.
siRNAs recruit DNA methylases and histone-modifying enzymes to targets
- Dicer-like (DCL) nuclease produces siRNAs which bind to Argonaute (AGO) proteins to bring modifying enzymes such as DRM1 and DRM2 to DNA targets.
- RNA-dependent RNA Polymerase (RdRP) amplifies the signal and produces secondary siRNAs
- Two plant-specific RNA polymerases, RNA Pol IV and RNA Pol V, contribute to siRNA-mediated silencing.
一、MIR Genes
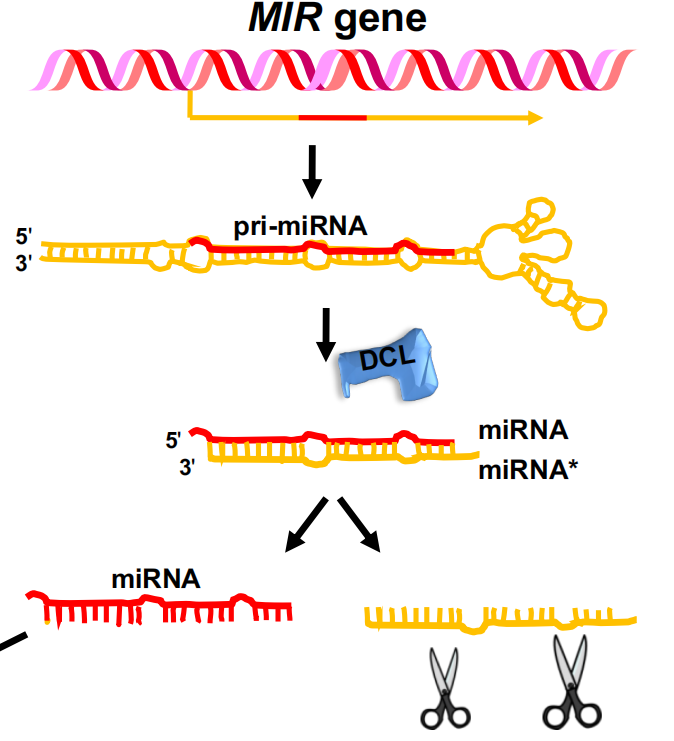
MIR genes are transcribed into long RNAs that are processed to miRNA
- miRNAs are encoded by MIR genes
- The primary miRNA (pri-miRNA) transcript folds back into a double-stranded structure, which is processed by DCL1
- The miRNA strand is degraded
Some MIR gene families are present in all plants or all angiosperms(被子植物) :
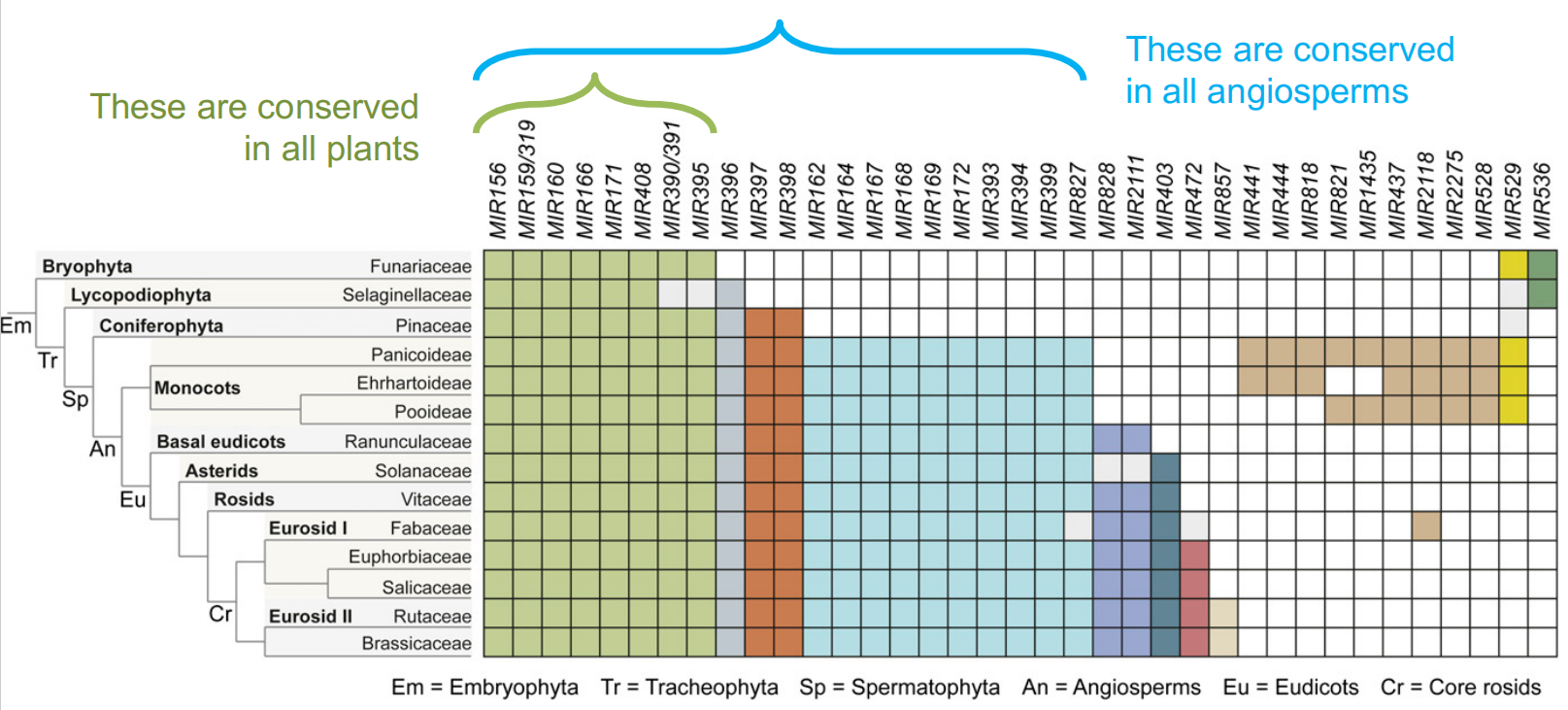
1. MIR156 – Highly Conserved
- miR156 is highly conserved within the plant kingdom
- miR156 is found in angiosperms as well as mosses (苔藓)
- miR156 is encoded by six or more genes in Arabidopsis
- miR156 targets transcription factors that control developmental phase changes
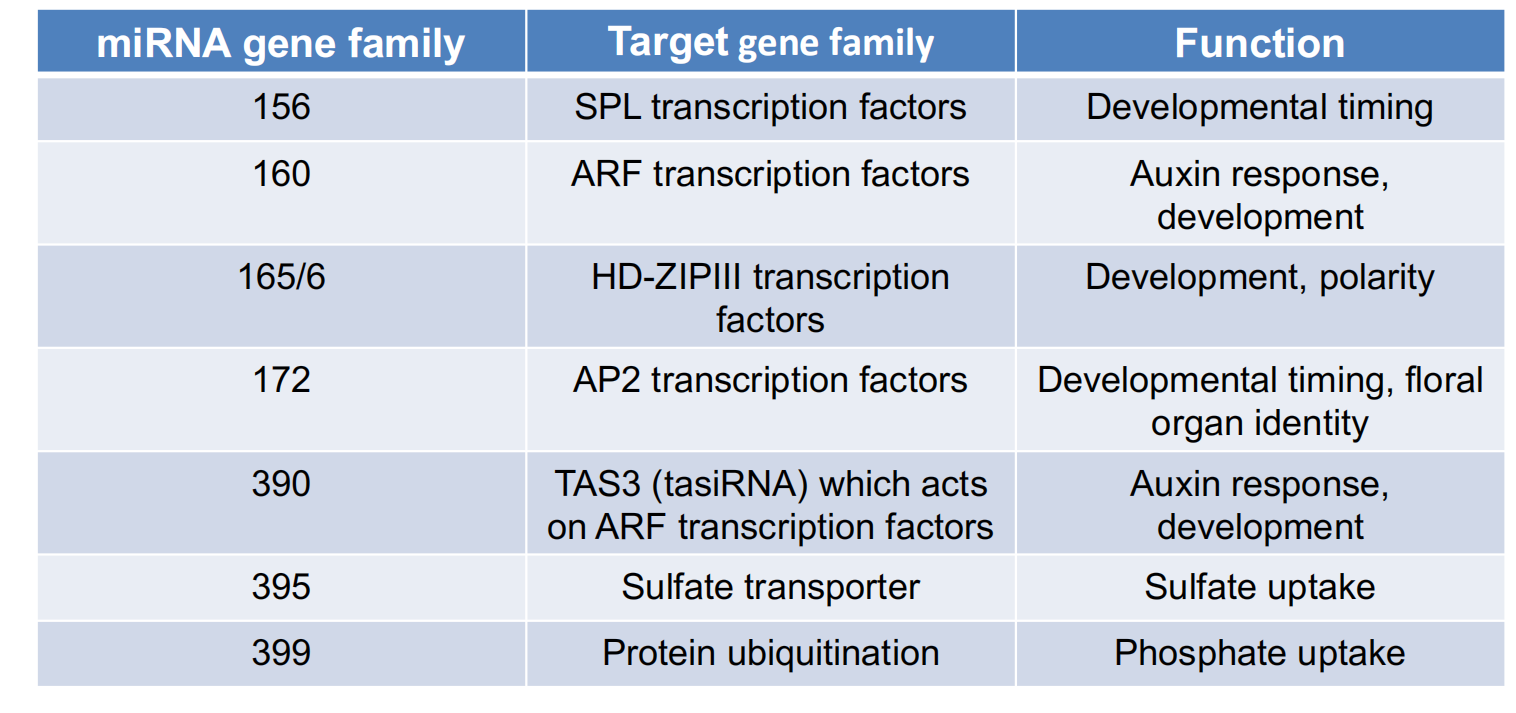
2. Targets of some conserved miRNAs
3. Plant miRNAs
Plant miRNAs are thought to be distantly related to their targets
Plant miRNAs are thought to be derived from their target sequences following gene duplication, inverted duplication and divergence.
Only some miRNAs confer selective advantage and are retained and further duplicated.
miRNAs and vegetative phase change
1. Phase Change
Vegetative phase change is the transition from juvenile to adult growth in plants.
Vegetative phase change affects morphology and reproductive competence
Phase change can affect leaf shape, phyllotaxy, and trichome patterns
Eucalyptus leaves are strongly dimorphic, as are leaves of holly and ivy. In other plants including Arabidopsis and maize the change is more subtle.
(桉树的叶子是强烈的二态的,就像冬青和常春藤的叶子一样。在包括拟南芥和玉米在内的其他植物中,这种变化更为微妙。)
In Arabidopsis, phase change affects leaf shape and trichome patterning
miR156 targets SPL genes, promoters of phase change
The function of miR156 in promoting developmental change is conserved
Phase change may involve a temporal cascade of miRNAs and transcription factor
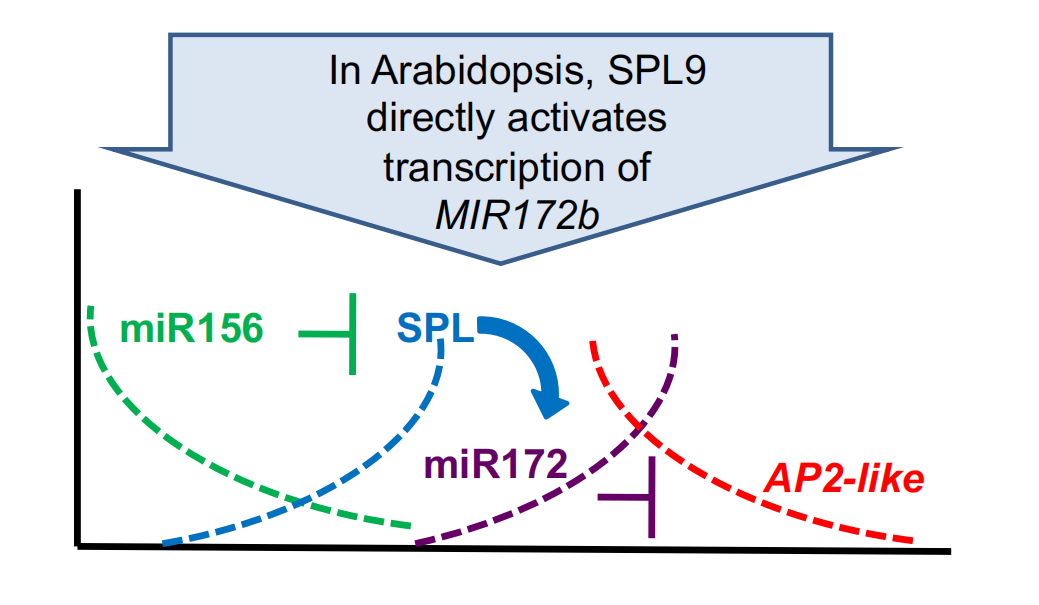
2. miR172
miR172 promotes flowering in Arabidopsis by targeting AP2-like transcription factors
miR172 expression temporally regulates AP2-like proteins
It is thought that floral initiation can occur when the level of AP2-like floral inhibitors drops below a certain level.
3. Summery
- Vegetative phase change affects morphology and reproductive compe
- miRNAs contribute to the temporal control of gene expression and phachange
- In the nematode C. elegans, lin-4 silencing of lin-14 is required for developmental progression
miRNAs contribute to developmental patterning
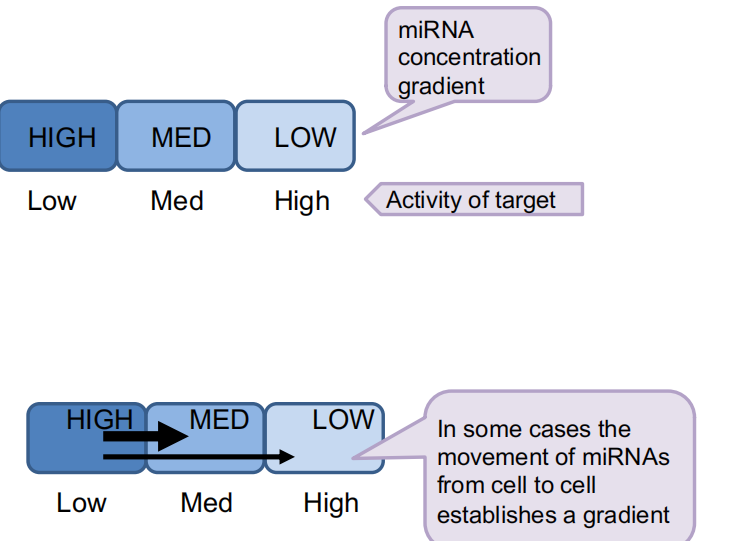
miRNA distribution patterns can spatially restrict activity of their targets
miRNAs can move between cells to spatially restrict activity of their targets
miRNAs and nutrient uptake – miR399
- The rate of nutrient uptake in the roots is regulated by signals from the shoot
- PHO2 encodes a ubiquitin-conjugating E2 that presumably contributes to directed proteolysis
- PHO2 levels are regulated by miR399 which accumulates upon phosphate starvation
- Starvation-induced miR399 moves from shoot to root, regulating phosphate uptake through PHO2
- PHO2 expression is also controlled by IPS1, a target mimic of miR399
三、The Small RNA World
tasiRNAs
1. Introduction
tasiRNAs = trans-acting siRNAS Encoded by TAS genes Primary transcript processing initiated by miRNA
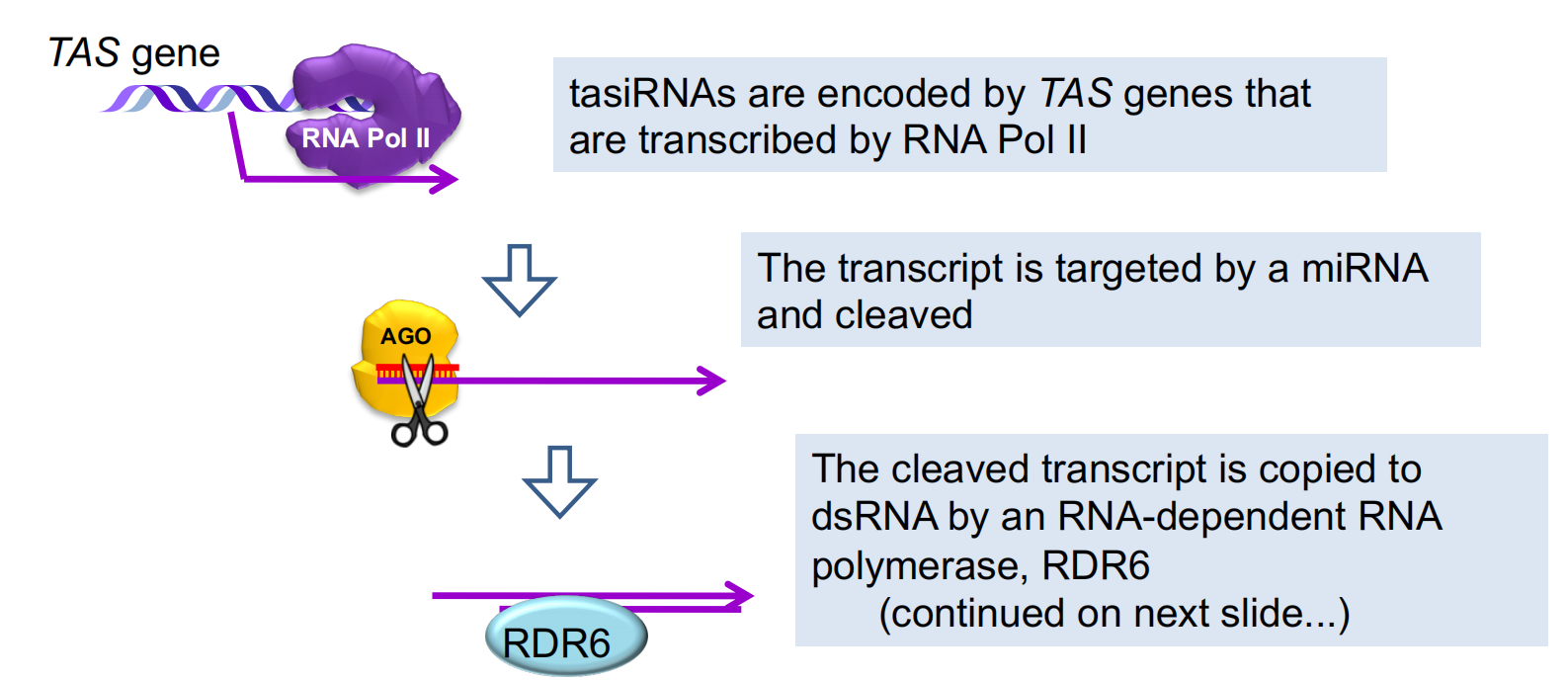
Arabidopsis has four families of TAS genes
TAS1 and TAS2 tasiRNAs target pentatricopeptide repeat genes.
TAS3 tasiRNAs target ARF transcription factors.
TAS4 tasiRNAs target MYB transcription factors
2. tasiRNA biogenesis
The dsRNA is cleaved by DCL4 into a series of shorter dsRNAs, releasing many tasiRNAs from a single TAS gene.
Arabidopsis
Arabidopsis has four families of TAS genes
TAS1 and TAS2 tasiRNAs target pentatricopeptide repeat genes.
TAS3 tasiRNAs target ARF transcription factors.
TAS4 tasiRNAs target MYB transcription factors
“phasiRNAs” – PHASED siRNAs
As part of a potent signal amplification system, these phased siRNAs function to post-transcriptionally silence genes, mainly in trans but also in cis.
Several “phased” tasiRNAs are derived from each TAS gene
Mutations that affect tasiRNA production affect phase change
四、The influence of RNA pol
Plants have additional RNA polymerase complexes that contribute to silencing
| Complex | Distribution | Function |
|---|---|---|
| RNA Polymerase I | All eukaryotes | Production of rRNA |
| RNA Polymerase II | All eukaryotes | Production of mRNA, microRNA |
| RNA Polymerase III | All eukaryotes | Production of tRNA, 5S rRNA |
| RNA Polymerase IV | Land plants | Production of siRNAs |
| RNA Polymerase V | Angiosperms | Recruitment of AGO to DNA |
Loss of function of RNA Pol IV interferes with silencing
Transcriptional silencing requires RNA Pol IV and V
RNA Pol IV contributes to siRNA production. Non-coding RNAs produced by RNA Pol V direct silencing machinery to target sites.
Loss of function of RNA Pol IV interferes with silencing
四、Transposons
Most siRNAs are produced from transposons and repetitive DNA
Most of the cellular siRNAs are derived from transposons and other repetitive sequences. In Arabidopsis, as shown above, there is a high density of these repeats in the pericentromeric (中心体周围) regions of the chromosome.
Epigenetic silencing of transposons and repetitive elements
Transposons must be tightly controlled to prevent widespread mutagenic activity. Epigenetic controls to maintain silencing include DNA methylation, histone modification and siRNA production.
五、DNA methylation
DNA can be covalently modified by cytosine methylation.
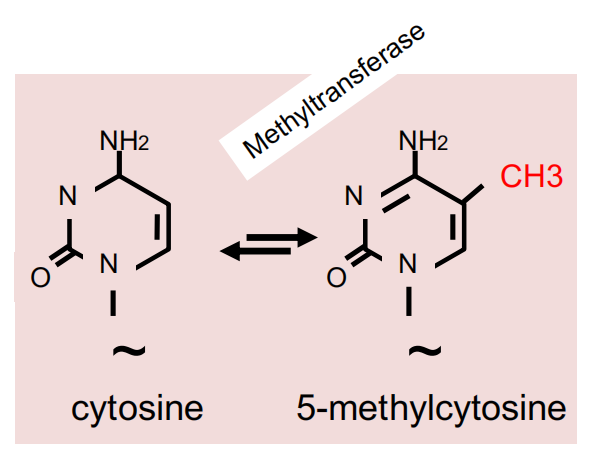
DNA methyltransferases in Arabidopsis
MET1 (METHYLTRANSFERASE1) – 5’-CG-3’ sites
- Silencing of transposons, repetitive elements, some imprinted gene
CMT3 (CHROMOMETHYLASE3) – 5’-CHG-3’ sites
- (H= A, C or T)
- Interacts with histone mark
- DRM 1, DRM 2 (DOMAINS REARRANGED 1 and 2) - 5’-CHH-3’ sites
- Primarily targets repetitive elements
- Requires the active targeting of siRNAs
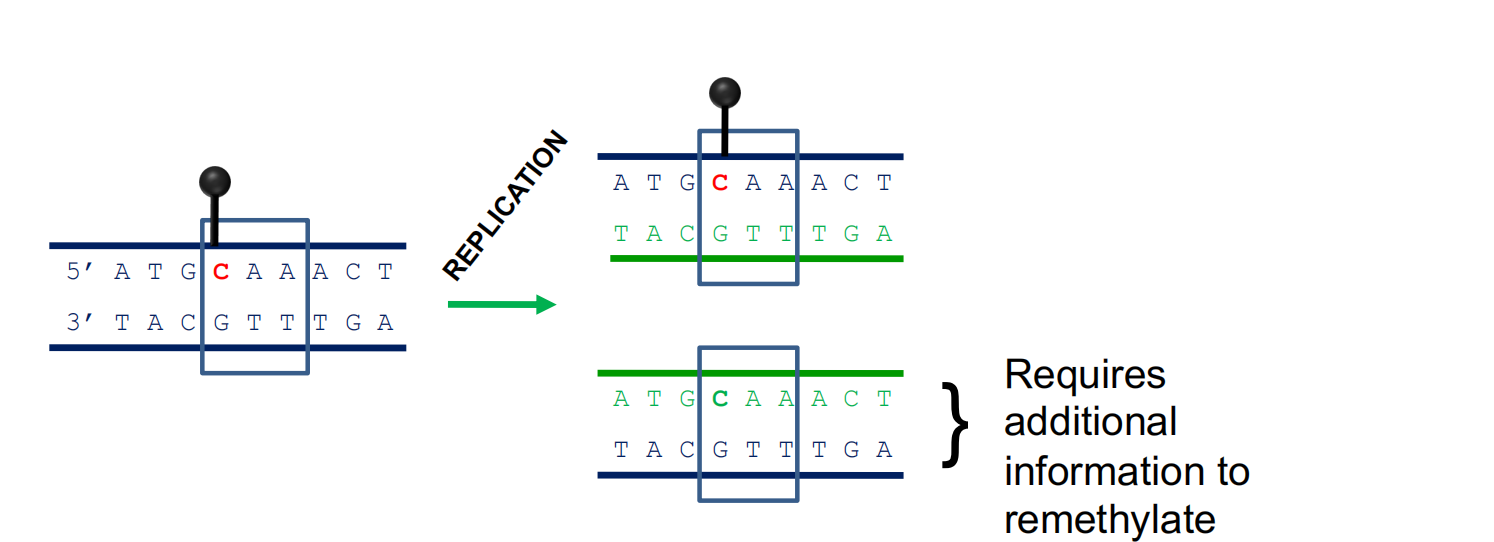
CG methylation can be propagated during DNA replication
1. Asymmetric Methylation
Asymmetric methylation sites require additional information
Asymmetric methylation sites are maintained (and initiated) by information on associated histones, and an RNA-based mechanism, RNA-directed DNA
Methylation (RdDM), that directs DNA methylases to these sites.
Some sites are maintained by small interfering RNAs (siRNAs)
Different DNA methylases act on different sites
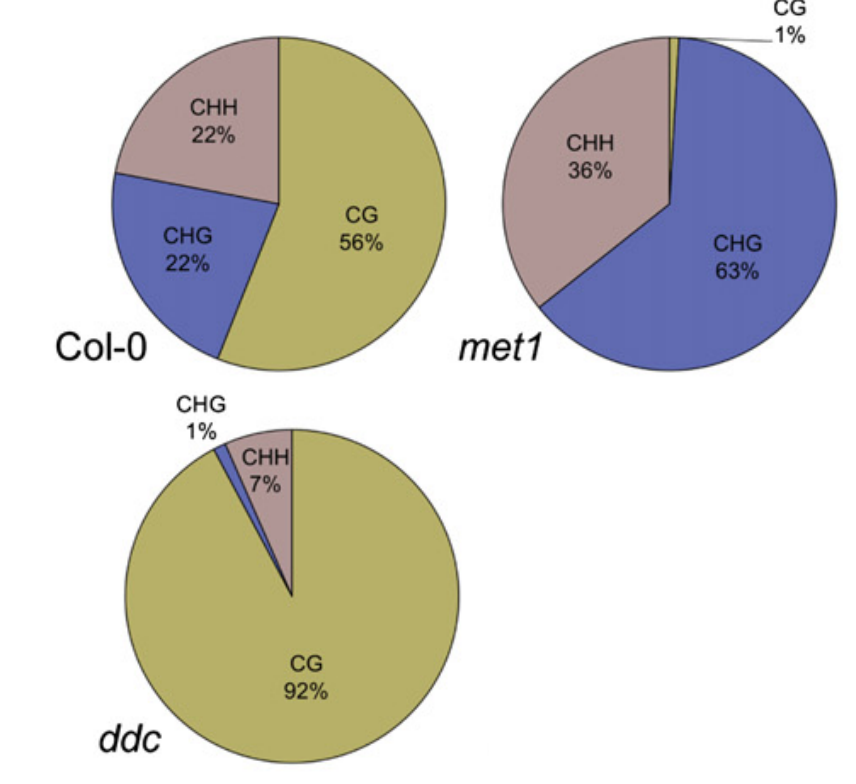
Heterochromatin DNA is highly methylated
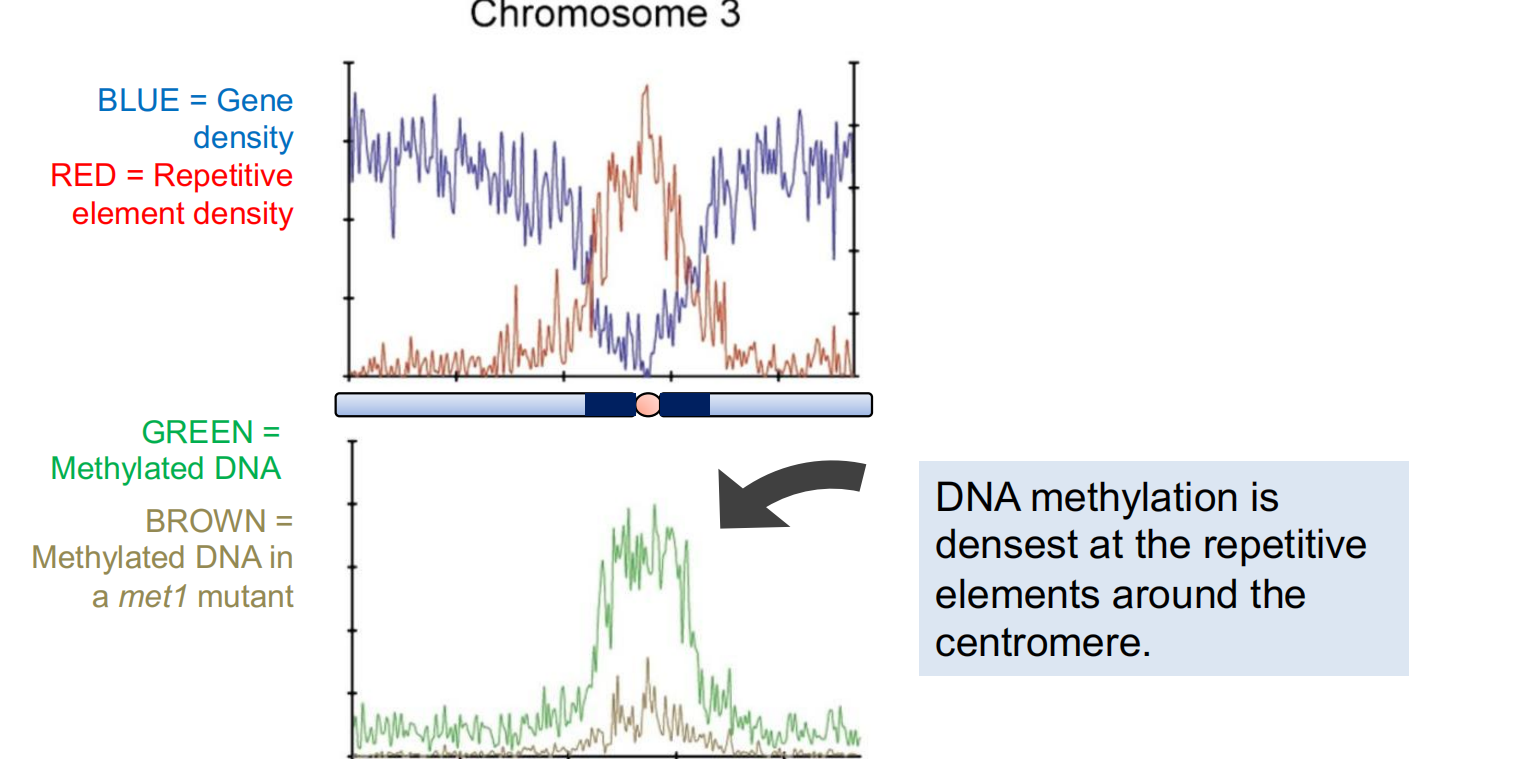
DNA methylation is densest at(最密集) the repetitive elements around the centromere.
Although CG methylation is more abundant in pericentromeric regions, a higher proportion of CHH methylation is found there.
六、siRNAs - summary
The siRNA pathway silences foreign DNA, transposons and repetitive elements.
In plants, siRNAs are produced by the action of Dicer-like proteins dicing dsRNA into 24 nt siRNAs
The siRNAs associate with AGO proteins and form silencing complexes
The silencing complexes can act post-transcriptionally on RNA targets, cleaving them or interfering with translation
The silencing complexes can also act on chromatin, silencing their targets by DNA methylation or histone modification