L02 The Structure of DNA & Chromosome
一、DNA
Central Dogma
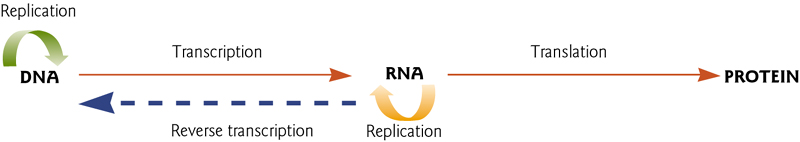
This defines the three important cellular processes:
- Replication of DNA
- Production of single-stranded RNA (“ssRNA”) from double-stranded DNA (dsDNA). This is known as “transcription”.
- Production of proteins from RNA. This is known as translation.
- Reverse transcription is rather unusual, but occurs mainly in viral-derived elements.
Components of Nucleic Acids
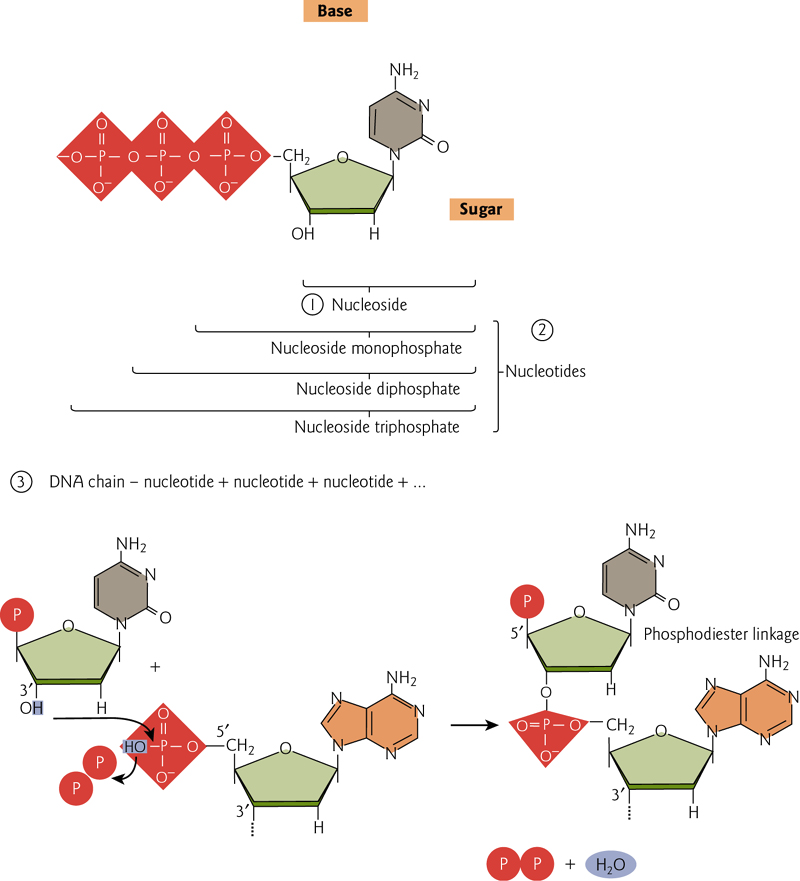
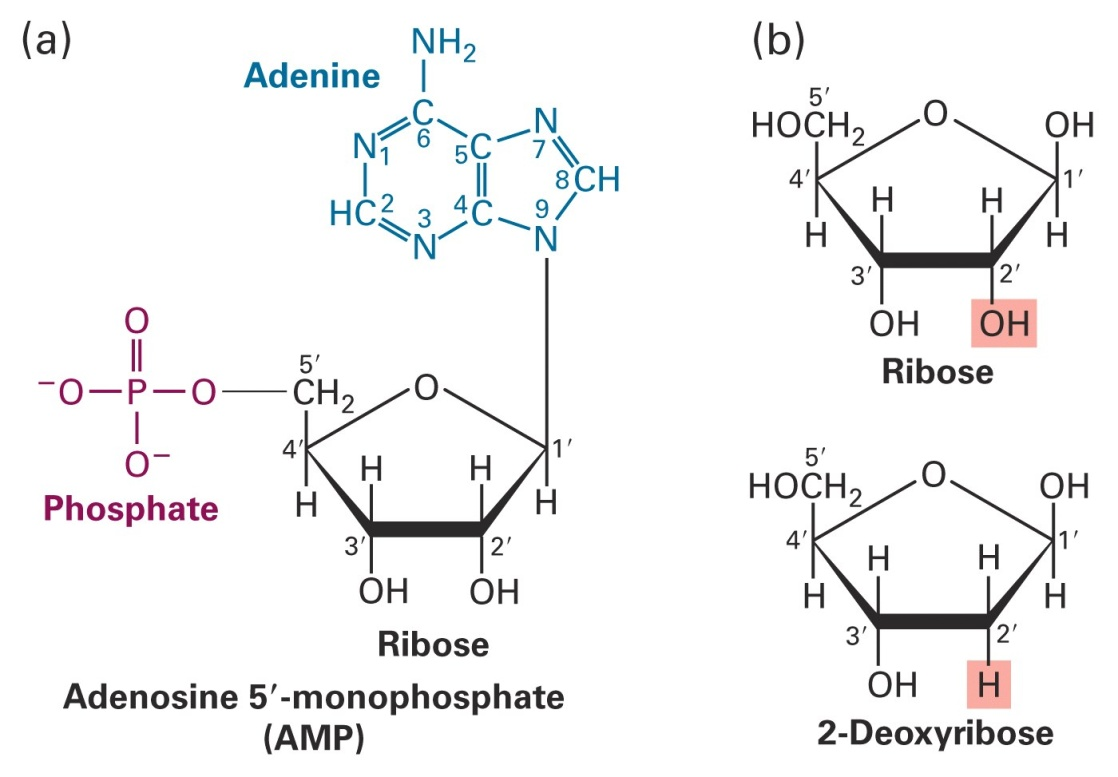
Nucleic acids, DNA and RNA, are comprised of monomeric units (nucleotides or “bases”) that have been joined together. These bases, sugars and phosphates are the fundamental building blocks
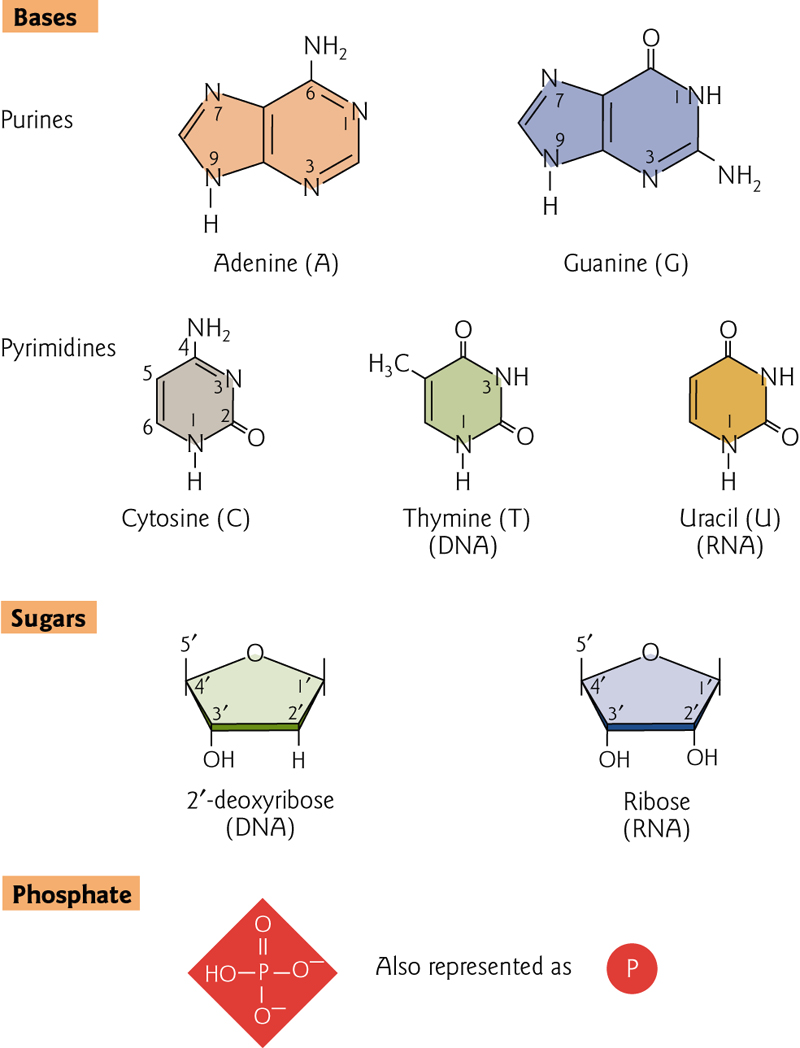
| Purine 嘌呤 | Pyrimidine 嘧啶 |
|---|---|
| Adenine(A)腺嘌呤 | Cytosine(C) 胞嘧啶 |
| Guanine(G)鸟嘌呤 | Thymine(T)胸腺嘧啶 |
| Uracil(U)尿嘧啶 |
The chemical nature of DNA
Monomeric units (nucleotides or “bases”) joined together to form a phosphodiester backbone with of repeating pentose-phosphate units.
Purine and pyrimidine bases extend as side groups
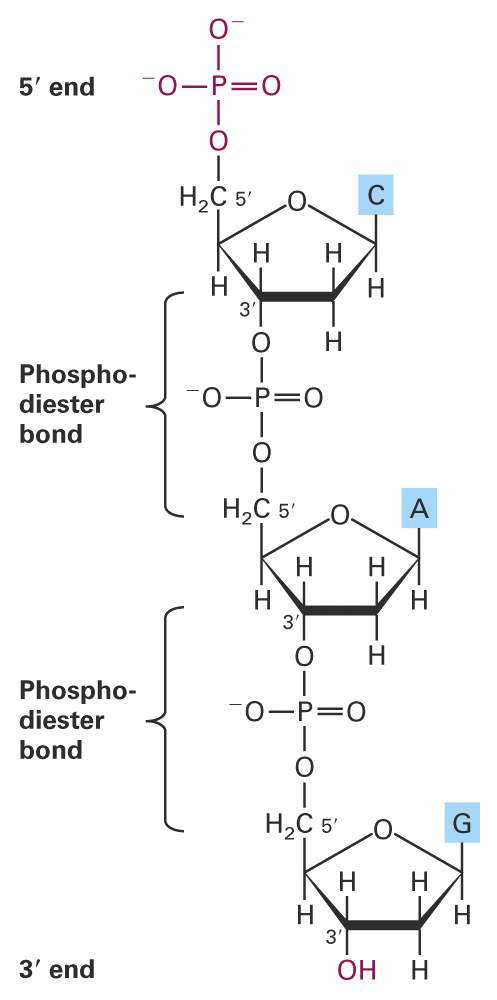
Like a polypeptide, a nucleic acid strand has an endto-end chemical orientation: the 5’ end has a hydroxyl or phosphate group on the 5’ carbon of its terminal sugar; the 3’ end usually has a hydroxyl group on the 3’ carbon of its terminal sugar.
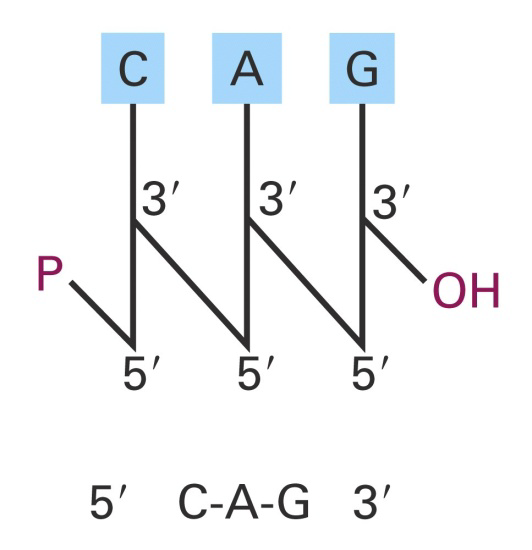
1. Base Composition in DNA
Chargoff
Examined base ratios from bacteria, animals, and plants. Whenever he determined the ratio of the two purines (adenine / guanine) the A:G ratio was found to vary dramatically. For example: 0.4 for bacteria versus 1.72 for yeast
Chargoff’s Rule states that [A] = [T] and [G] = [C]
The ratio of purines to pyrimidines was always 1:1. Out of these observations the idea of base pairing between purines and pyrimidines emerged.
The G-C content is definded as:
$$
\frac{G+C}{A+T+G+C}
$$
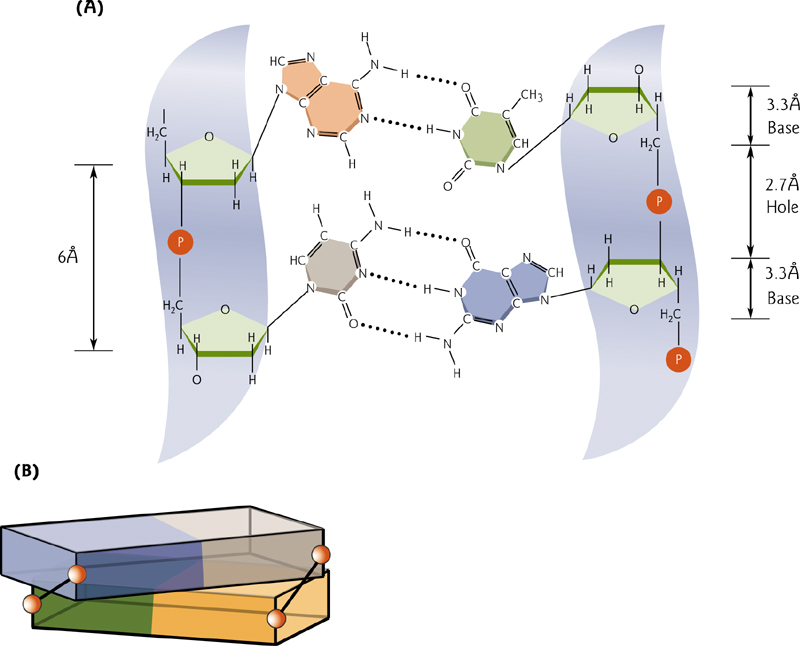
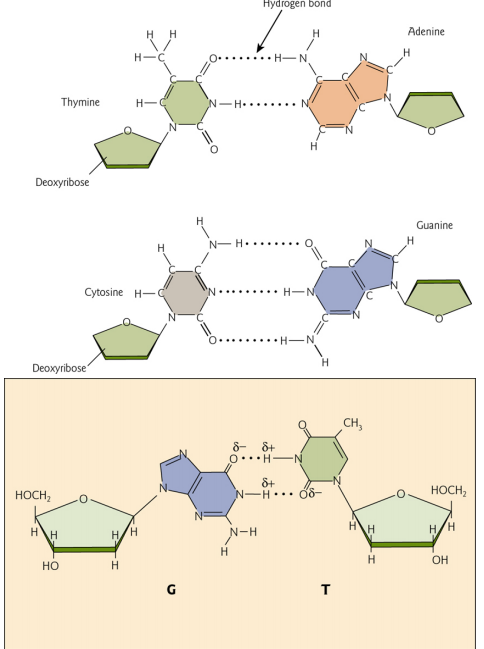
G-C and A-T pairing
Guanine-cytosine pairs use three hydrogen bonds, adenine-thymine use two.
2. Nucleic acids are comprised of nucleotides
Each nucleotide consists of a cyclic 5-carbon sugar, a purine or pyrimidine base attached to the sugar, and a phosphate.
3. G-C and A-T pairing
Guanine-cytosine pairs use three hydrogen bonds, adenine-thymine use two.
Structure of DNA
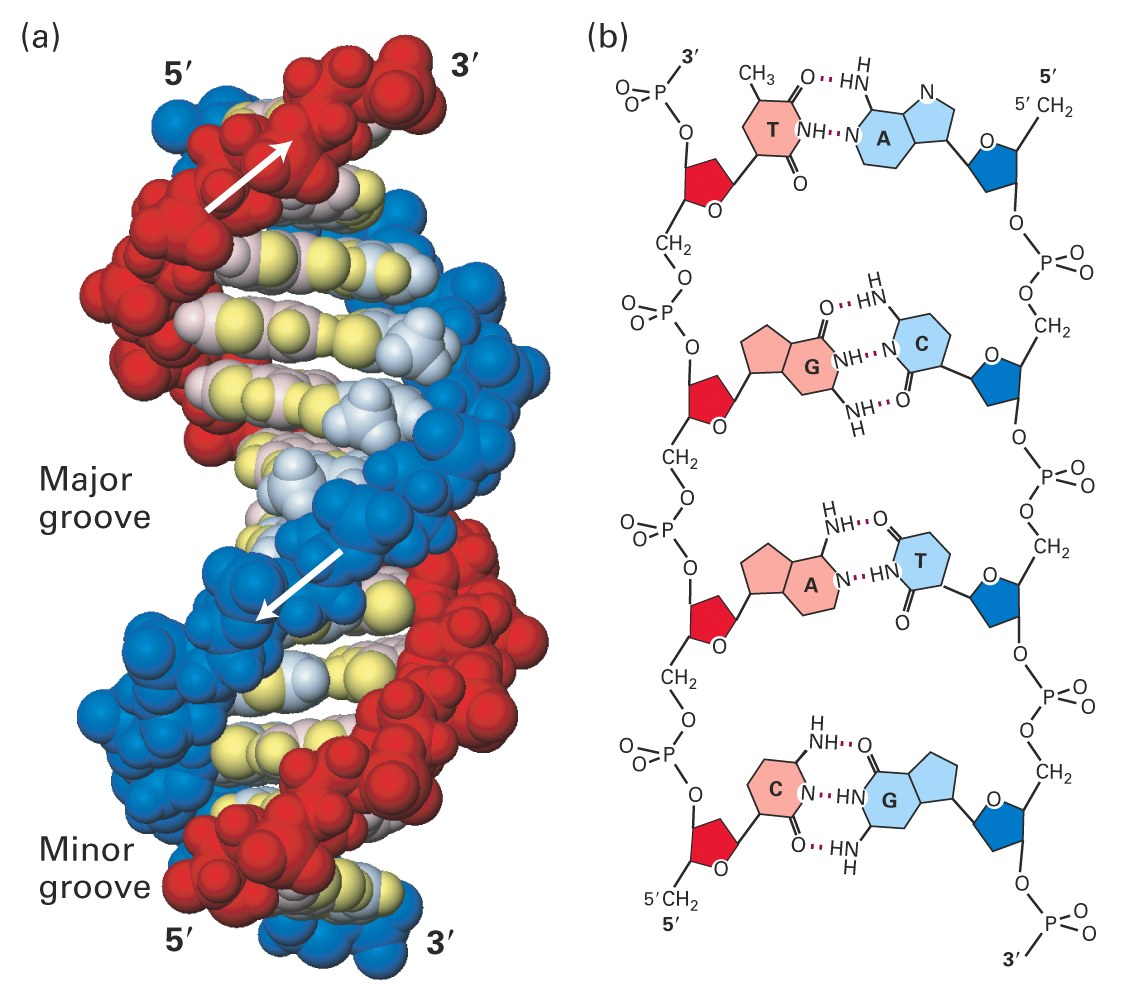
1. DNA: comprised of anti-parallel strands held together by hydrogen bonds.
This is an important concept: the “top” and “bottom” strands of DNA are in opposite directions and are complementary(互补的) (A pairs with T and G pairs with C).
By convention (and because synthesis proceeds in this direction), the 5’ end of the top strand is drawn at the left:
This is another way to draw two strands of DNA, but it means the same as a more complicated cartoon like this:
2. DNA structures
Three DNA structures have been described.
In low humidity(湿度), B DNA changes to the A form; RNA-DNA and RNA-RNA helices exist in this form.
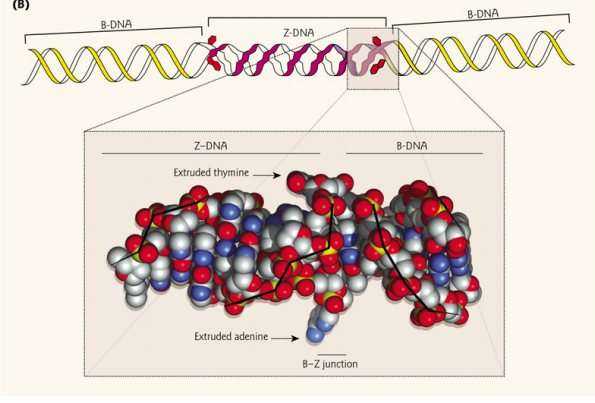
Short DNA molecules can adopt an alternative left-handed configuration instead of the normal right-handed helix called Z DNA because the bases seem to zigzag when viewed from the side.
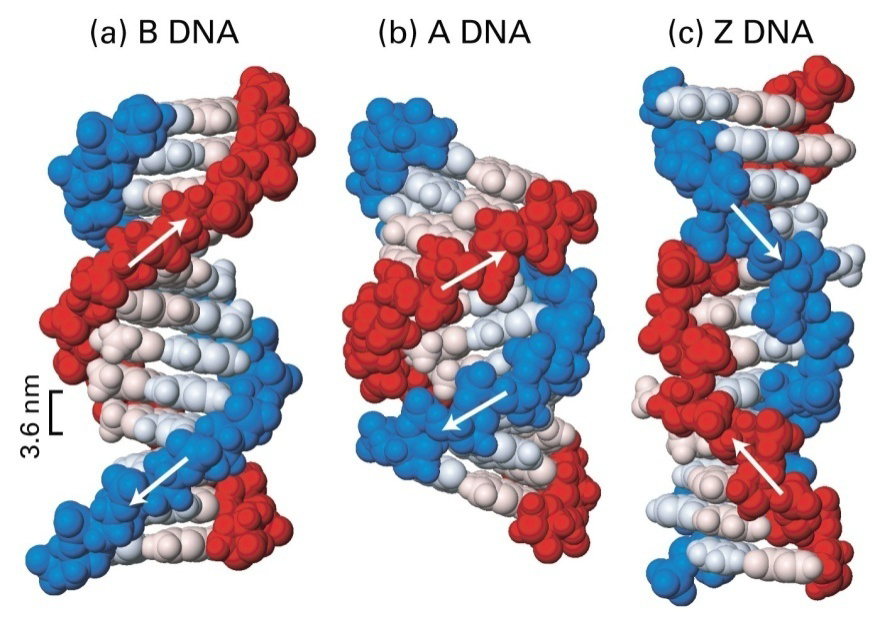
DNA denaturation(变性) and hybridization(杂交)
Denatured macromolecules have disrupted 3-D structures, compared to the ‘natural’ state. DNA can be disrupted by heating or high pH (basic) treatment to break the hydrogen bonds between the two strands.
DNA can be disrupted by heating or high pH (basic) treatment to break the hydrogen bonds between the two strands.
Denaturation of DNA
Thermal stability of DNA molecules – helix favored by stacked bases and hydrogen bonding by anti-parallel strands.
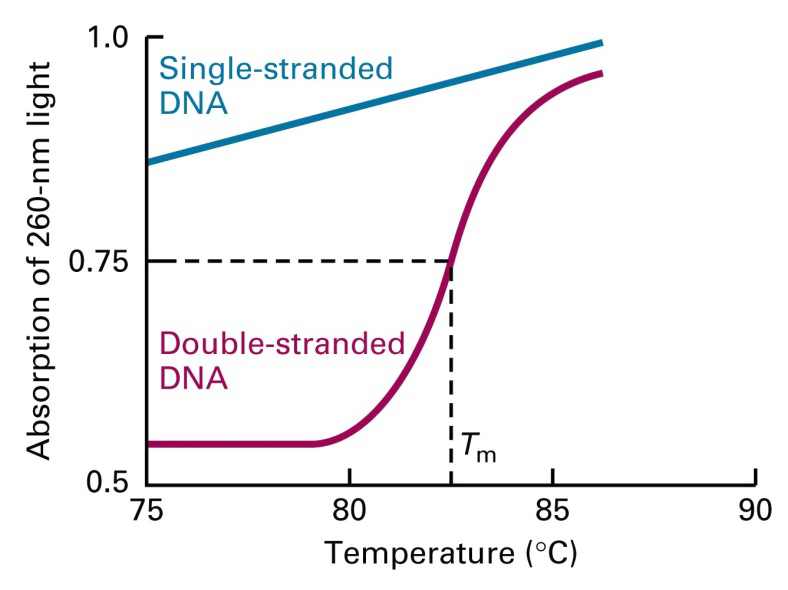
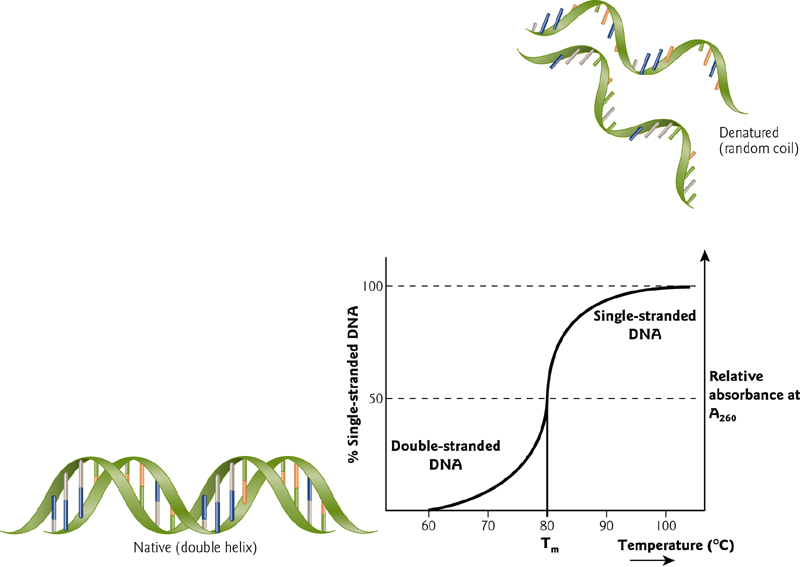
It is possible to follow denaturation in a spectrophotometer, by observing the change in absorbance at 260nm. Unstacked bases (random orientation) absorb more light than neatly stacked (oriented) base-pairs. This results in a melting curve. The rise in $A_{260}$ takes place rapidly.
Stability of DNA & Bases of Molecular tools and techniques
The polar charges in each strand of the DNA attract, thus making double stranded more stable than single stranded.
Therefore, if you optimize the conditions, it is possible to get only specific hybridization between nearly identical fragments. Less optimal conditions produce mis-paired bases
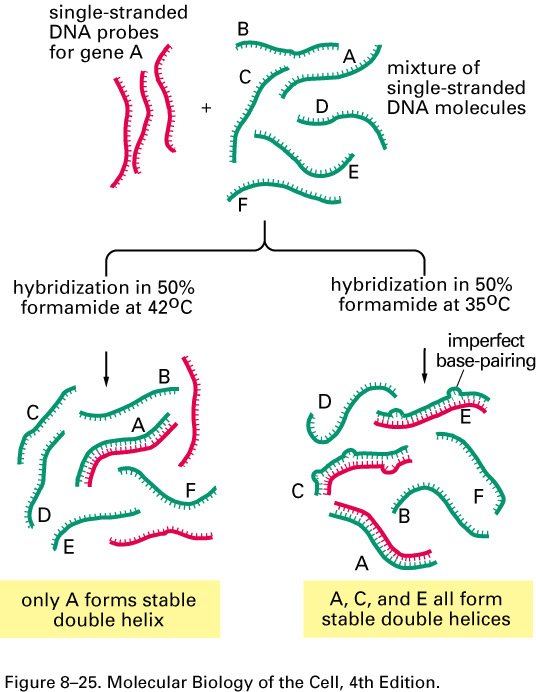
1. Tm (Melting Temperature)
Tm = melting temperature (rise in A260 is half complete)
The polar charges in each strand of the DNA attract, thus making double stranded more stable than single stranded.
Therefore, if you optimize the conditions, it is possible to get only specific hybridization between nearly identical fragments. Less optimal conditions produce mis-paired bases
2. G–C Bond and Tm
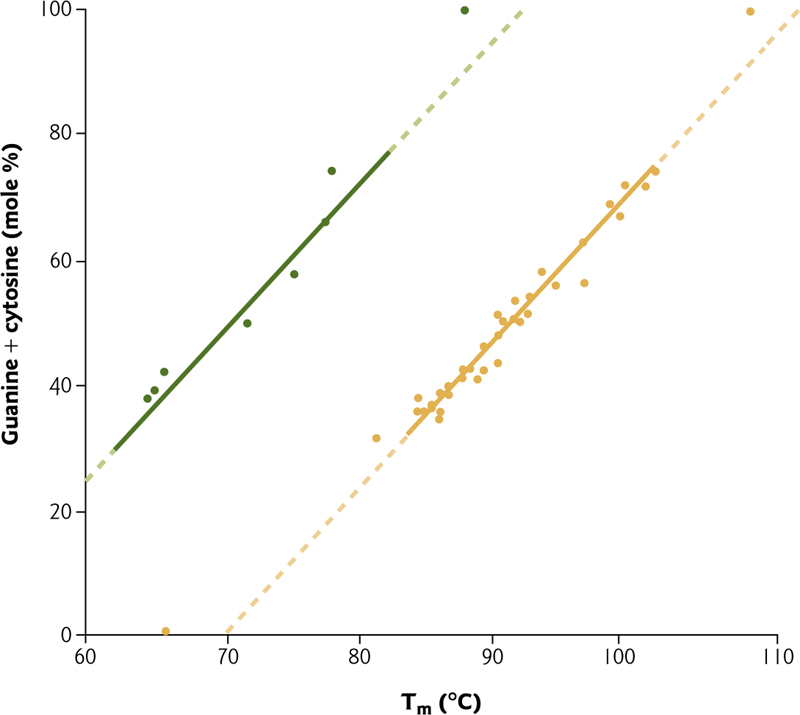
This indicates that G-C pairs are more stable than A-T pairs – which we know to be true because guanine and cytosine form THREE hydrogen bonds, while adenine and thymine form TWO hydrogen bonds.
3. Renaturation of DNA (aka “reannealing)
An important concept – many molecular tools are based on this, because only very similar strands of DNA will reanneal. If you can mark one of the strands (radioactively, for example), it allows you to “find” a homologous strand.
Requirements for renaturation:
- High salt to eliminate electrostatic interference of phosphates.
- High temperature to break random hydrogen bonds (20 to 25°C below Tm)
Renaturation is a random process by which a strand finds its complementary partner.
Renaturation is a random process by which a strand finds its complementary partner.
As with denaturation, the change in $A_{260}$ is used to monitor the status of the DNA in solution
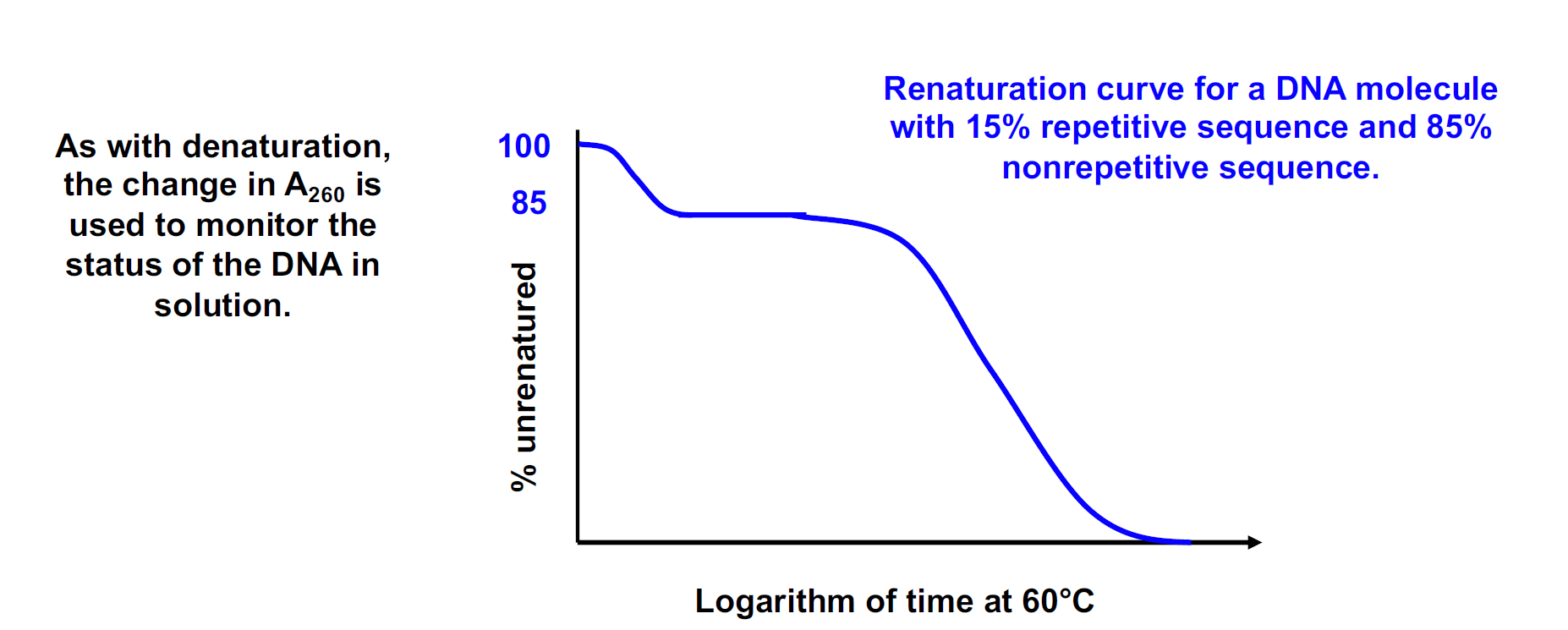
As with denaturation, the change in A260 is used to monitor the status of the DNA in solution.
DNA secondary structures
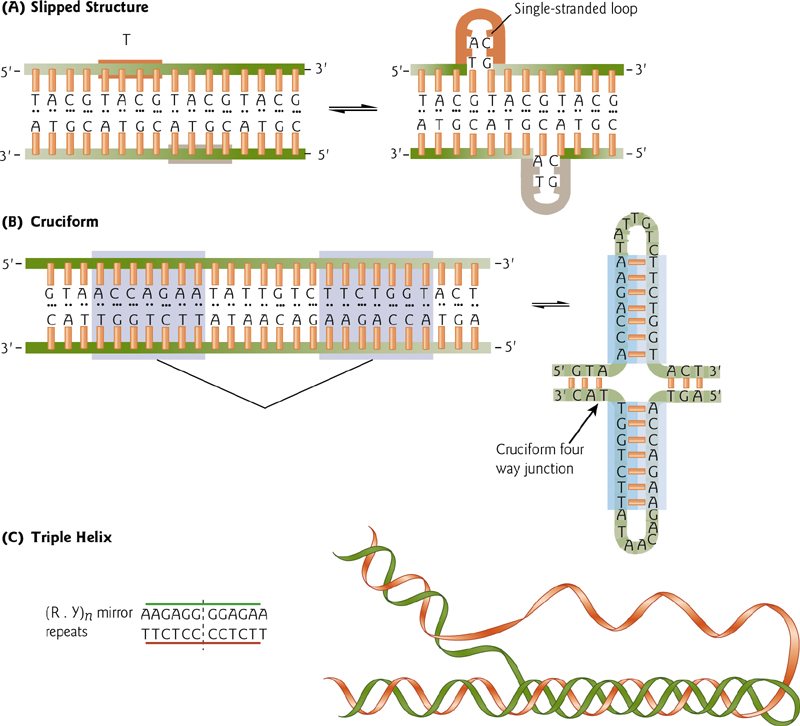
Patterns in DNA
1. Tandem repeats 串联重复
Tandem repeats – multiple copies of a short sequence, perhaps 2 to 5 bp, but can be much longer.
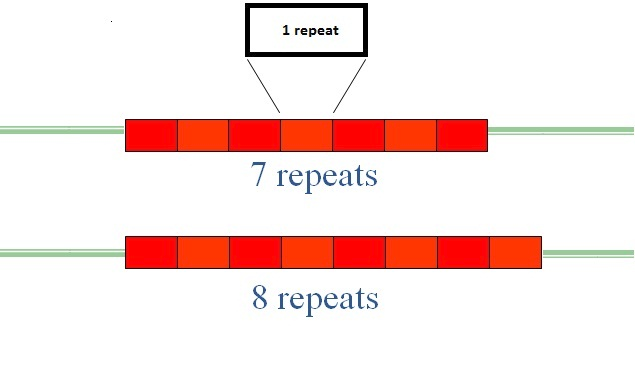
由于串联重复在不同个体中有着特异性,因此可以用来进行亲缘关系识别
2. Inverted Repeats 反向重复序列
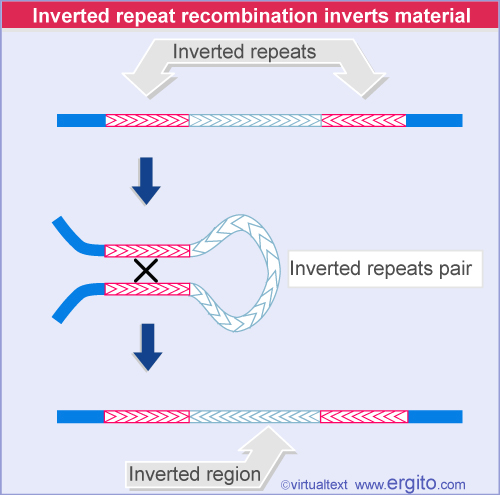
a pair of similar or identical sequences, in an inverted orientation. Because of base-pairing, these can fold back on themselves ultimately can facilitate recombination.
DNA supercoiling 超螺旋
脱氧核糖核酸链在双螺旋基础上如绳索般扭转的现象与过程称为DNA超螺旋。当脱氧核糖核酸处于“松弛”状态时,双螺旋的两股通常会延着中轴,以每10.4个碱基对旋转一圈的方式扭转。但如果脱氧核糖核酸受到扭转,其两股的缠绕方式将变得更紧或更松。当脱氧核糖核酸扭转方向与双股螺旋的旋转方向相同时,称为正超螺旋,此时碱基将更加紧密地结合。反之若扭转方向与双股螺旋相反,则称为负超螺旋,碱基之间的结合度会降低。
T = number of turns of DNA
L = number of times one strand crosses the other.
L < T is a negative supercoil
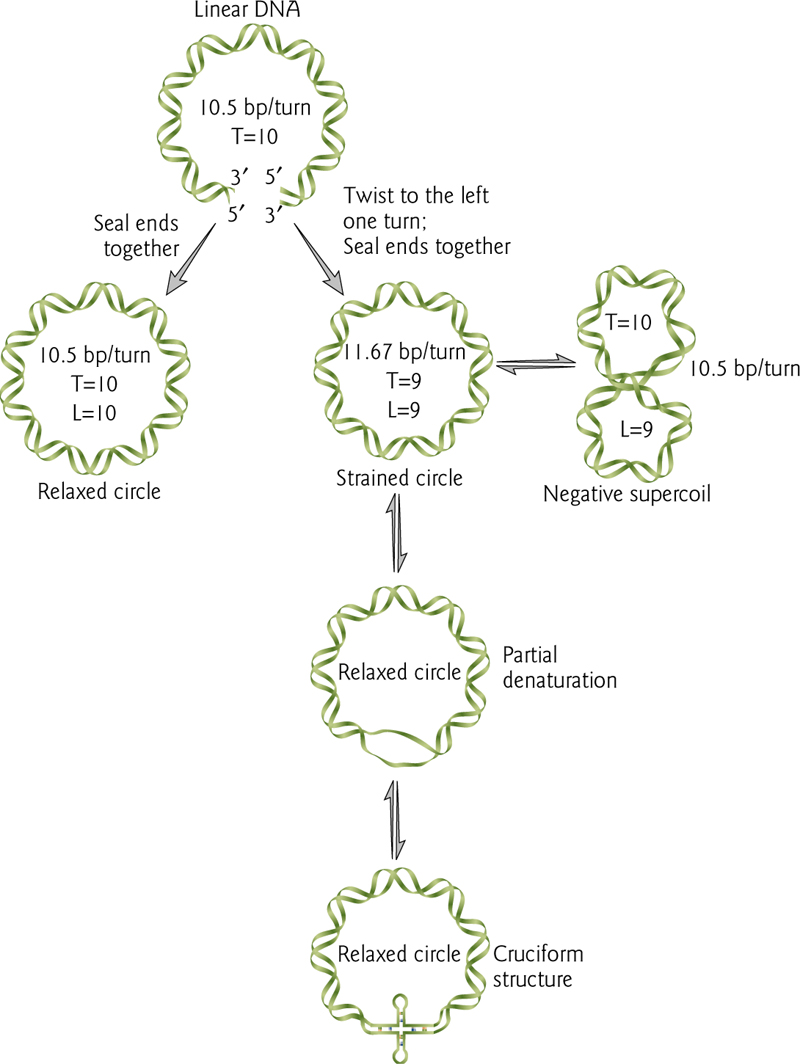
1. Topoisomerase
自然界中大多数的脱氧核糖核酸,会因为拓扑异构酶的作用,而形成轻微的负超螺旋状态。拓扑异构酶同时也在转录作用或DNA复制过程中,负责纾解脱氧核糖核酸链所受的扭转压力。
Topoisomerase nicks one strand allowing the other to rotate, either introducing or releasing additional supercoils.
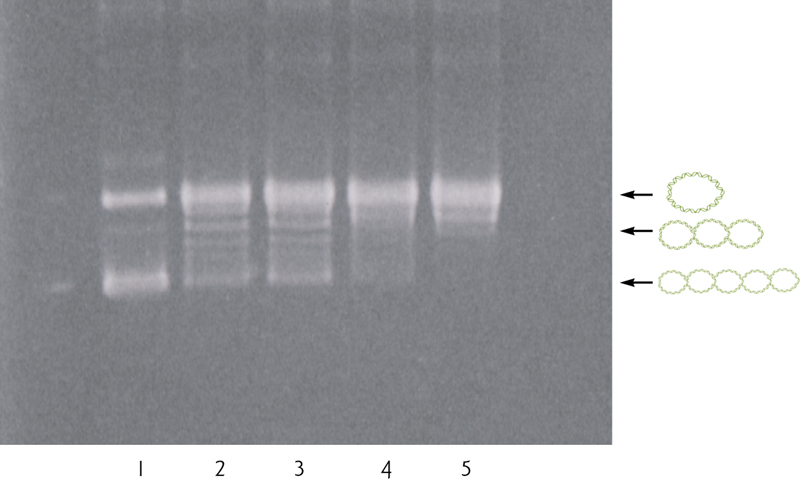
Macromolecule isolation and characterization: DNA electrophoresis
Electrophoresis: separation of nucleic acids or proteins-on the basis of size, electric charge, and other physical properties.
This usually takes place in a gel, under the influence of an electric field.
二、Genome Organization: from nucleotides to chromatin
Genome Size Varies
Classical measurements of DNA content showed that genome size is highly variable among diverse organisms
Eukaryotic genome sizes vary enormously, unrelated to intuitive ideas of morphological complexity (与形态复杂性的直观概念无关)
In prokaryotes, genome size and gene number are strongly correlated, but in eukaryotes the vast majority of nuclear DNA is non-coding.
The genome size may not reflect the number of genes, Hence the largest genomes are less density filled with genes.
Genome size are approximate, even when the genome is complete because of gaps in the sequence.
The length of DNA is vastly larger than the cell that contains it.
Genome Packing
The length of DNA is vastly larger than the cell that contains it
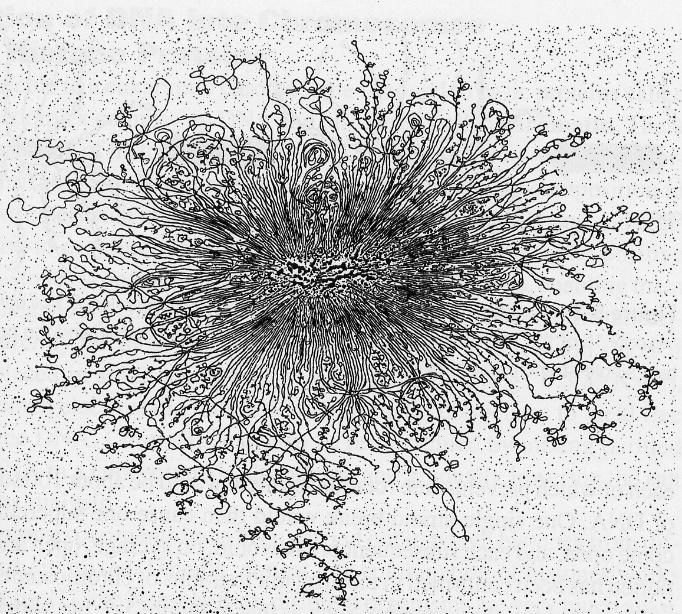
1. E.coli Genome
The E. coli chromosome is 1,300 um in length, but the cell is only 1x3 um.
The DNA forms a tightly supercoiled “nucleoid” structure with a dense protein-containing “scaffold” core.
The degree of supercoiling can be determined by centrifugation; nicked DNA is less dense, and one nick will release one loop of the DNA.
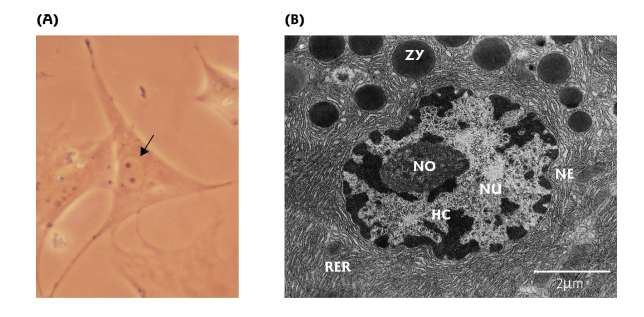
2. Eukaryotic Genome
The eukaryotic genome is located in the cell nucleus
What’s In a Genome?
Microscopically visible chromosomal parts:
- Heterochromatin
- Euchromatin
- Centromere
- Telomere
- Nucleolar organizing region
Sequence defined parts
- Genes (exons/introns)
- Protein coding and non-coding (miRNAs)
- rRNAs, tRNAs, snRNAs
- Regulatory sequences
- Promoters, enhancers, repressors, insulators
- Transposons and retrotransposons
- Small heterochromatic regions
1. Microscopically visible chromosomal parts
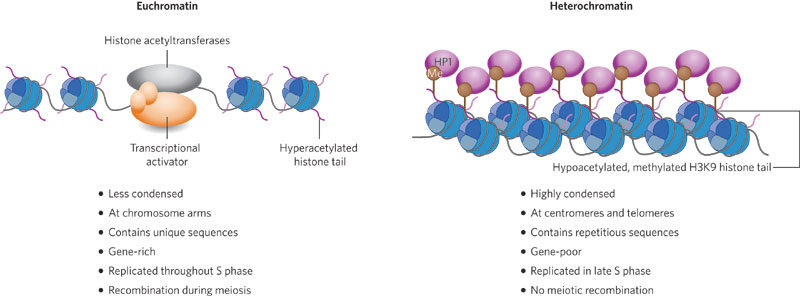
Heterochromatin 异染色质
During interphase, tightly coiled, heavily stained chromosomal material. Repetitive and genetically inactive. Define by small RNAs, in plants?
异染色质大多是由不具遗传活性的卫星串行(satellite sequences)所构成。其中的基因皆受到不同程度抑制,有些即使在真染色质内也无法表现。
- less condensed
- At chromosome arms
- Contains unique sequences
- Gene-rich
- Replicated throughout S phase
- Recombination during meiosis 减数分裂时重组
Euchromatin 常染色质
genetically active, lightly stained regions. Gene rich.
又译同染色质或常染色质,是基因密度较高的染色质,多在细胞周期的S期进行复制,且通常具有转录活性,能够生产蛋白质。真染色质在真核生物与原核生物的细胞中皆存在。与其相对而言,另一类通常无法转录成为蛋白质的染色质,则称为异染色质(heterochromatin)。在细胞中,92%的人类基因体是真染色质,剩余部分是异染色质。
- Highly condensed
- At centromeres and telomeres
- Contains repetitious sequences
- Gene-poor
- Replicated in late S phase
- No meiotic recombination (减数分裂重组)
Centromere 着丝粒
A highly condensed and constricted region of a chromosome. During mitosis, spindle fibers attach to this point to direct segregation of chromosomes.
染色体着丝粒(centromere),又称中节,主要作用是使复制的染色体在有丝分裂和减数分裂中可均等地分配到子细胞中。
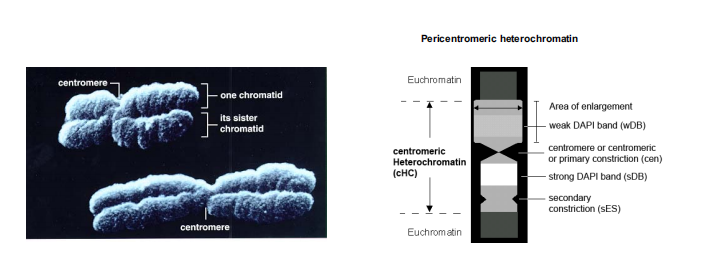
Telomere 端粒
repetitive structures at the end of the chromosome that function to protect chromosome ends during replication.
端粒(英语:Telomere)是染色体末端的DNA重复串行,作用是保持染色体的完整性和控制细胞分裂周期。
Nucleolar organizing region 核仁组织区
核仁组织区(Nucleolus Organizing Region,简称NOR)具有组织核仁的能力。人的13、14、15、21、22号染色体上有核仁组织区。
2. Sequence defined parts
Genes (exons/introns)
内含子是阻断基因线性表达的序列。DNA上的内含子会被转录到前体RNA中,但RNA上的内含子会在RNA离开细胞核进行转译前被剪除。在成熟mRNA被保留下来的基因部分被称为外显子。真核生物的基因含有外显子和内含子,是前者区别原核生物的特征之一。
(1) Protein coding and non-coding (miRNAs 小分子核糖核酸)
miRNA通过与目标信使核糖核酸(mRNA)结合,进而抑制转录后的基因表达,在调控基因表达、细胞周期、生物体发育时序等方面起重要作用。在动物中,一个微RNA通常可以调控数十个基因。
(2) rRNAs, tRNAs, snRNAs
Regulatory sequences
(1) Promoters, enhancers, repressors, insulators
启动子,增强子,抑制子,绝缘子
Transposons and retrotransposons
转座子(英语:Transposon,亦称为转座元件,跳跃子)是一类DNA序列,它们能够在基因组中通过转录或逆转录,在内切酶(Nuclease)的作用下,在其他基因座上出现。转座子的这种行为,与假基因(Pseudogene)的出现颇有相似甚至相同之处。有些科学家将后者视为“基因化石”,是透视物种进化的痕迹之一[1]。转座子的发现,证明了基因组并不是一个静态的集合,而是一个不断在改变自身构成的动态有机体。根据转座子“跳跃”方式的不同,转座子被分为I型(复制–粘贴)和II型转座子(剪切–粘贴)。
Small heterochromatic regions
小型异染色质区域,染色时比常染色质的颜色要深
During interphase, tightly coiled, heavily stained chromosomal material. Repetitive and genetically inactive.
Higher order structure and regulatory regions of eukaryotic chromosomes
形成更高级结构的方式就是Packaging(包装)
1. Condensed Chromosomes
Chromosomes condense during cell division, and condensed chromosome equals many levels of packaging
The long linear eukaryotic chromosomes are dramatically condensed: approximately 2 meters of DNA fits in each cell.
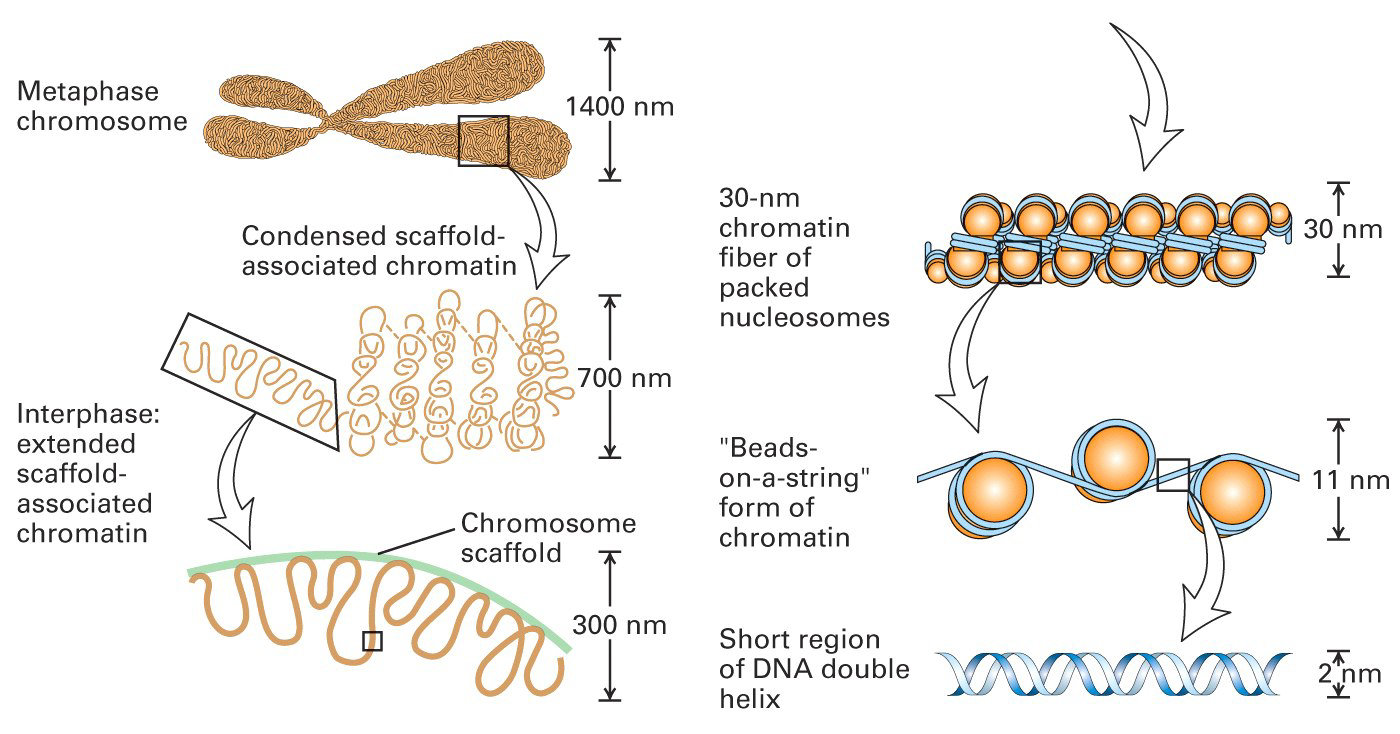
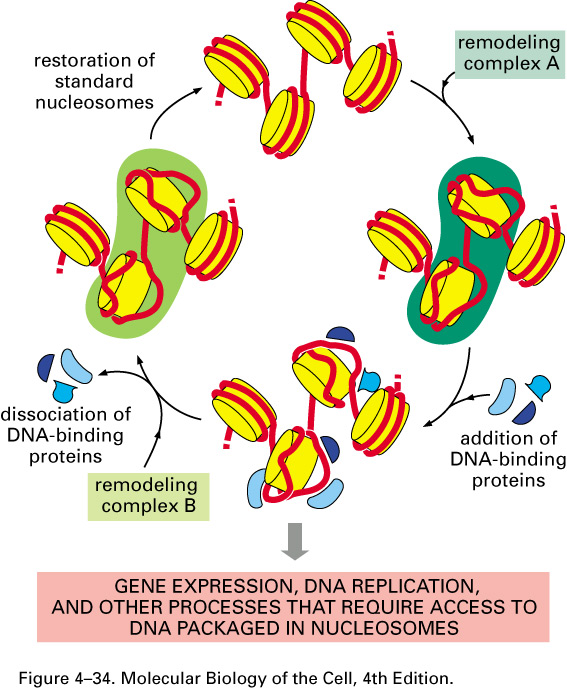
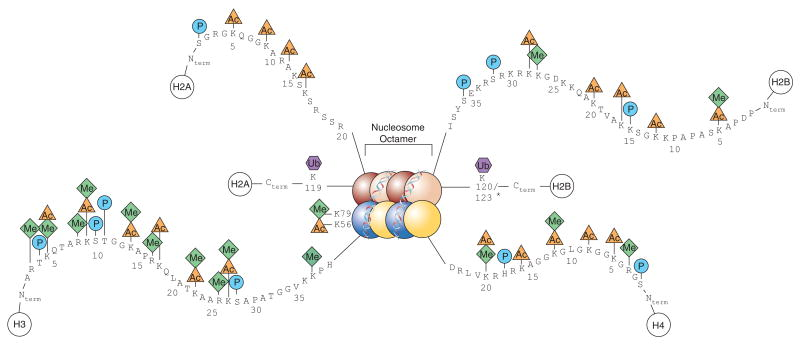
2. Nucleosome 核小体
Nucleosomes are the basic units of eukaryotic chromosome structure.
Two types of proteins are bound to the DNA which are classified as histones and non-histone chromosomal proteins.
The complex of both classes of proteins with nuclear DNA is known as chromatin
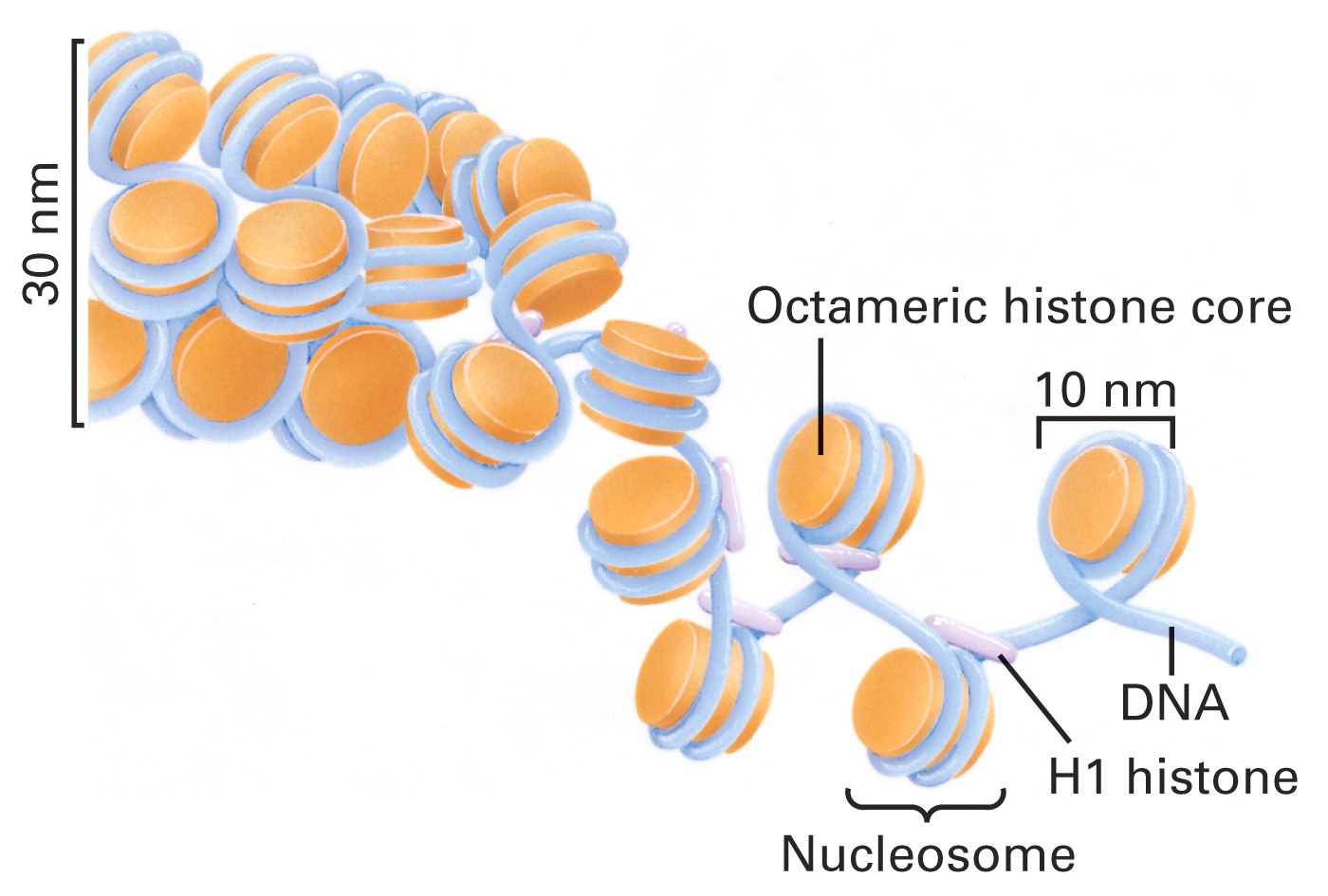
Partial digestion of chromatin produces ~180 bp bands, while more complete digestion produces 147 bp bands. 表明一个核小体包含的DNA长度为147 bp.
- Core histones H3, H2B, H2A, H4
- Linker histones H1, H5
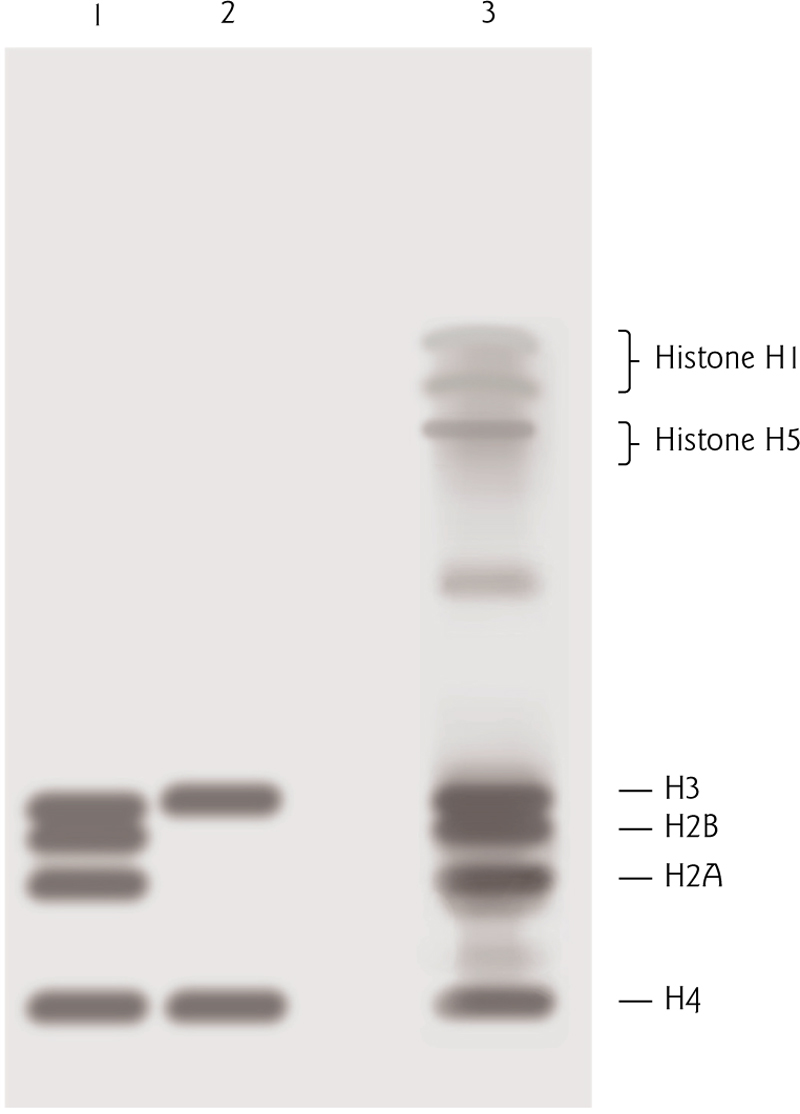
This is a “nonspecific” interaction – the specific sequence of the DNA does not affect this interaction.
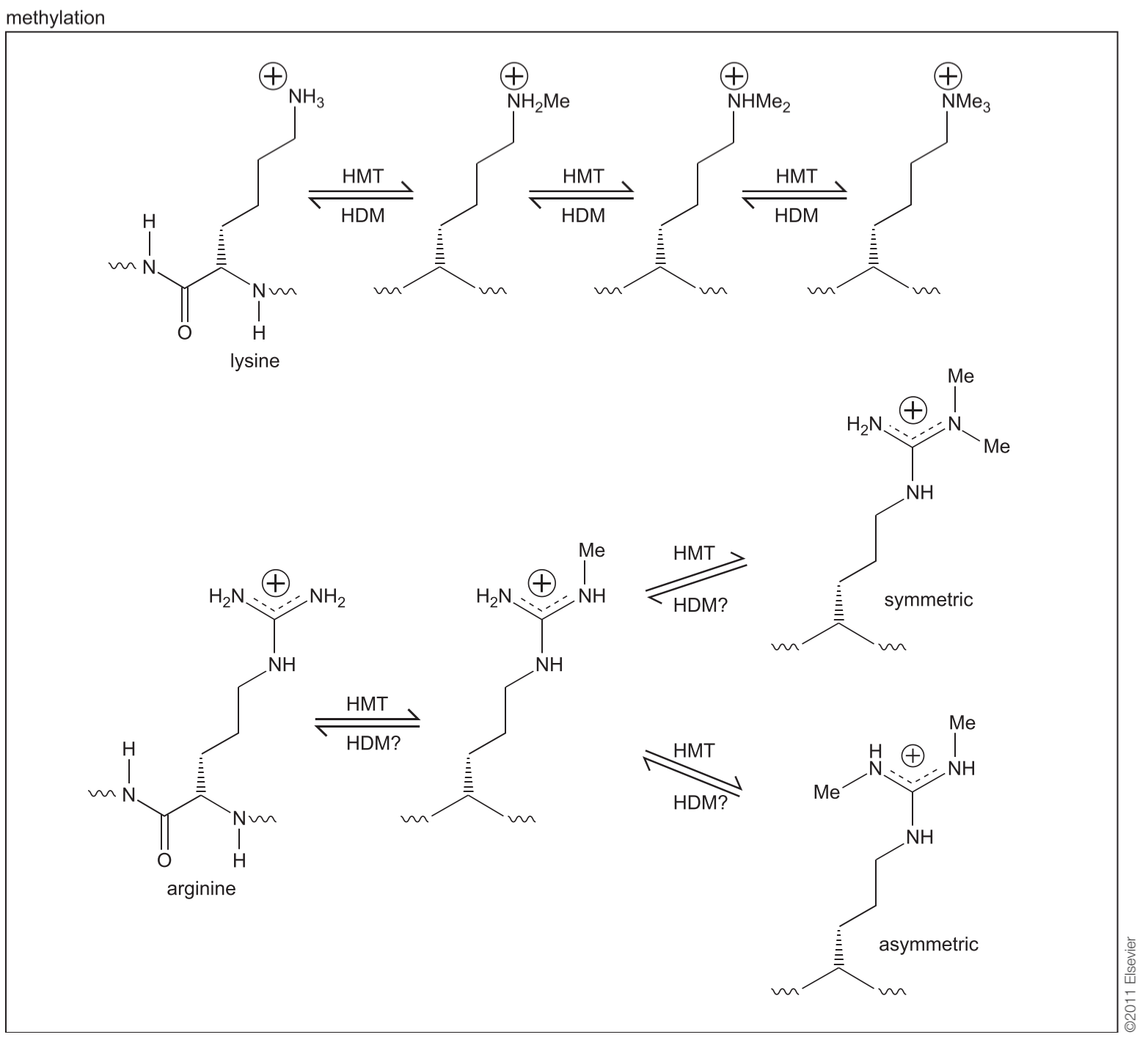
3. Histone Modification
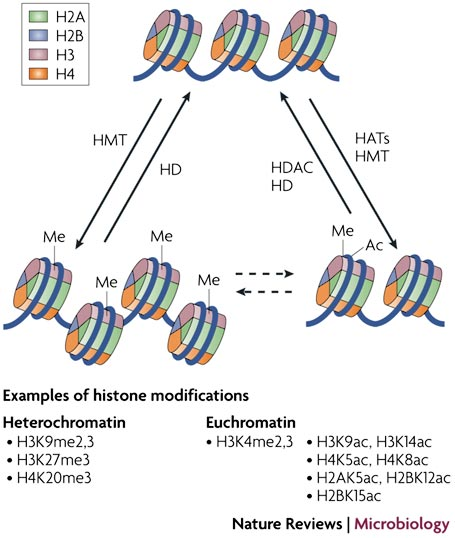
Relationship Between Heterochromatin And Euchromatin
The two states of chromatin are reversible:
- Heterochromatin - “closed” form that is associated with a silenced state.
- Euchromatin - “open” or active chromatin which allows gene expression.
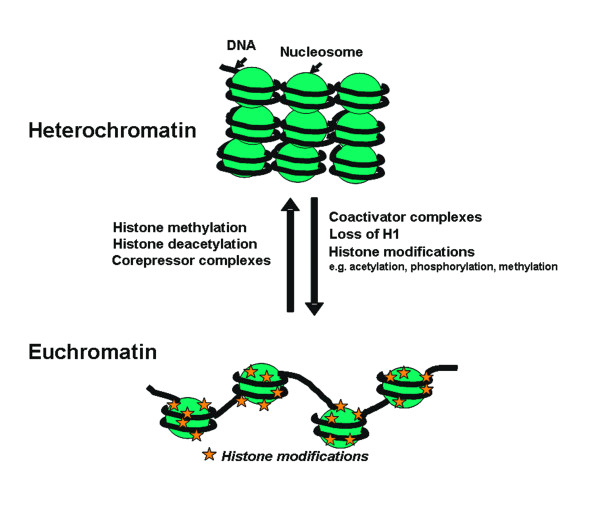
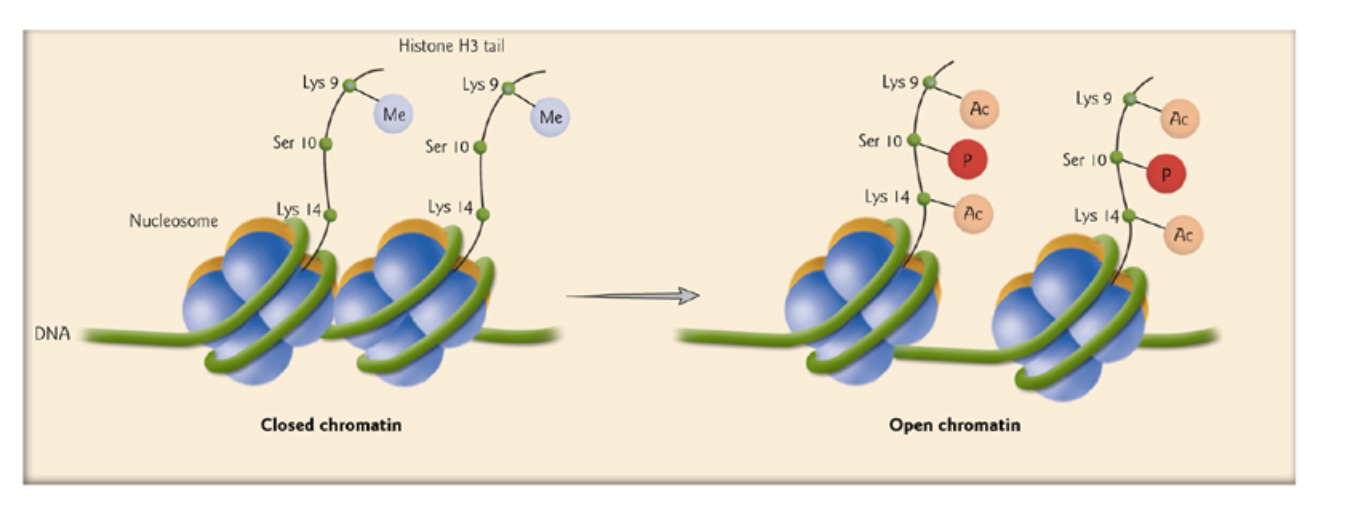
Histone modification alter nucleosome function
Post-translational modifications of the core histone tails that stick out from the nucleosomes have been directly linked to the regulation of chromatin structure, a concept known as the histone code
从核小体中伸出的核心组蛋白尾部的翻译后修饰(Post-translational modifications)与染色质结构的调控直接相关,这一概念被称为组蛋白密码(histone code)
Different types of proteins recognize and bind modified histones, and these modifications can regulate the availability of the DNA to different activities.
Depending on diverse histone modifications, a variety of multi-protein complexes wilbe recruited, thus allowing for tightly regulated transcriptional control.
One obvious change is that acetylation(乙酰化作用) and phosphorylation(磷酸化作用) each acts to reduce the overall positive charge of the histone tails
(1) acetylation
Acetyl groups are added to histone tails, by HAT’s (Histone acetyl transferases).
Acetyl groups are taken off by Histone deacetyl transferases (HDAC’s)
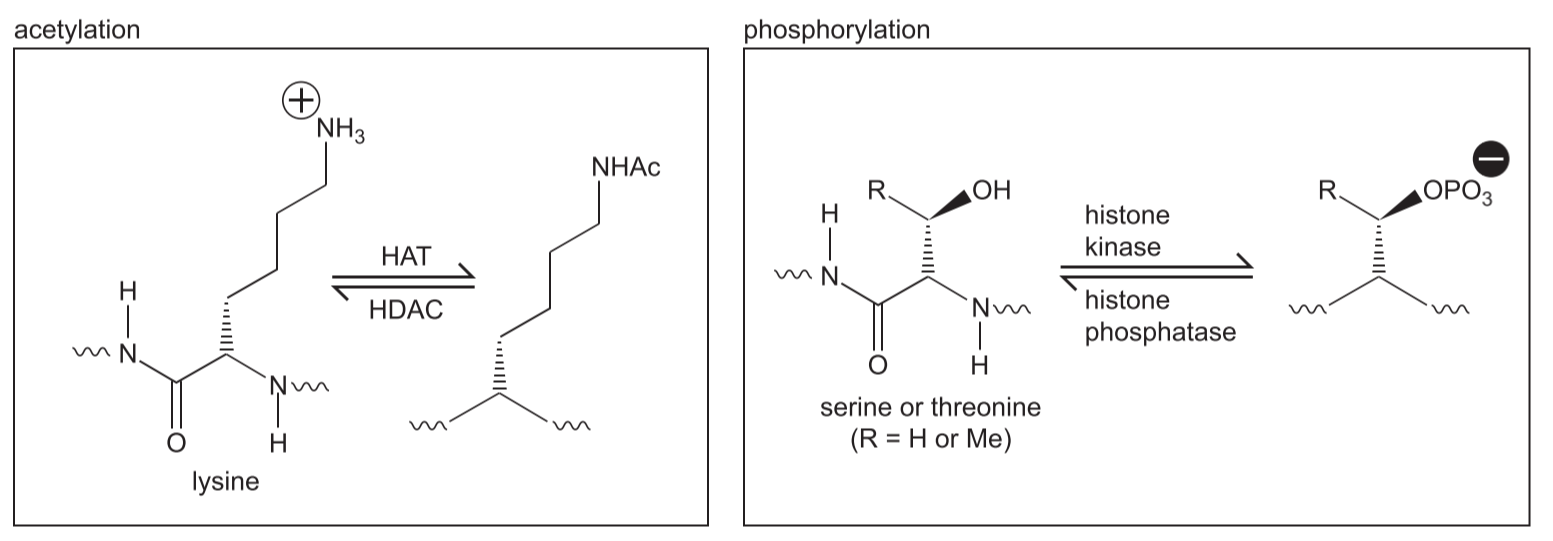
Acetylation of histone tails, carried out by histone acetyltransferases(乙酰转移酶), is primarily associated with active gene expression. 乙酰化使得异染色质向常染色质转化,而脱乙酰化与甲基化使得常染色质转化为异染色质
Histone acetylation results in the relaxation of the basic chromatin structure through increased charge repulsion and by serving as binding sites for protein complexes of chromatin-modifying and transcriptional activators.
Acetylation of lysine(赖氨酸) neutralizes its positive charge (Fig. 8-41). This loss of positive charge reduces the affinity of the tails for the negatively charged backbone of the DNA.
Diversity of histone modification
Depending on diverse histone modifications, a variety of multi-protein complexes will be recruited, thus allowing for tightly regulated transcriptional control.
Ways to Study The Histone Modification
It’s possible to use “chromatin immunoprecipitation” (Chromatin-IP, or ChIP) to identify DNA sequences associated with specific histone modifications
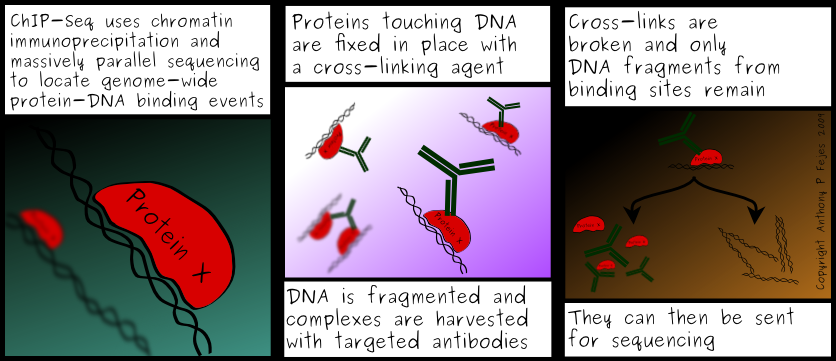
三、Different Genome
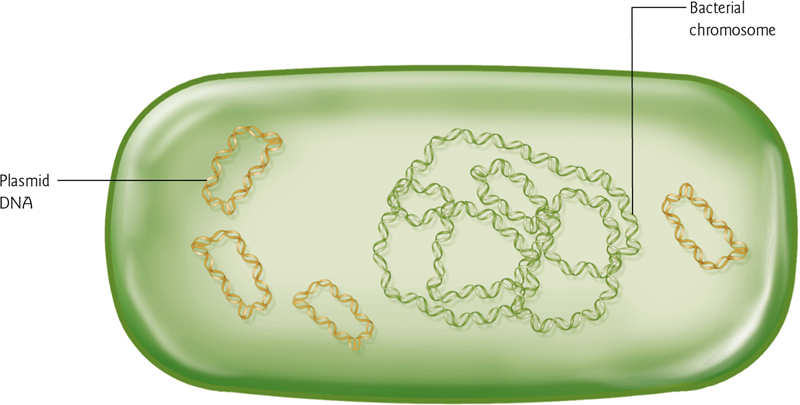
Bacterial Genome
No histones in bacteria! So no heterochromatin or euchromatin.
Bacterium also contain circular plasmids – extrachromosomal & self-replicating. These are important tools for molecular biology.
Organelle DNA
Majority of eukaryotic DNA is in nucleus but some is present in mitochondria and chloroplast
Organelle DNA is like the bacterial genome: without nucleus, circular DNA
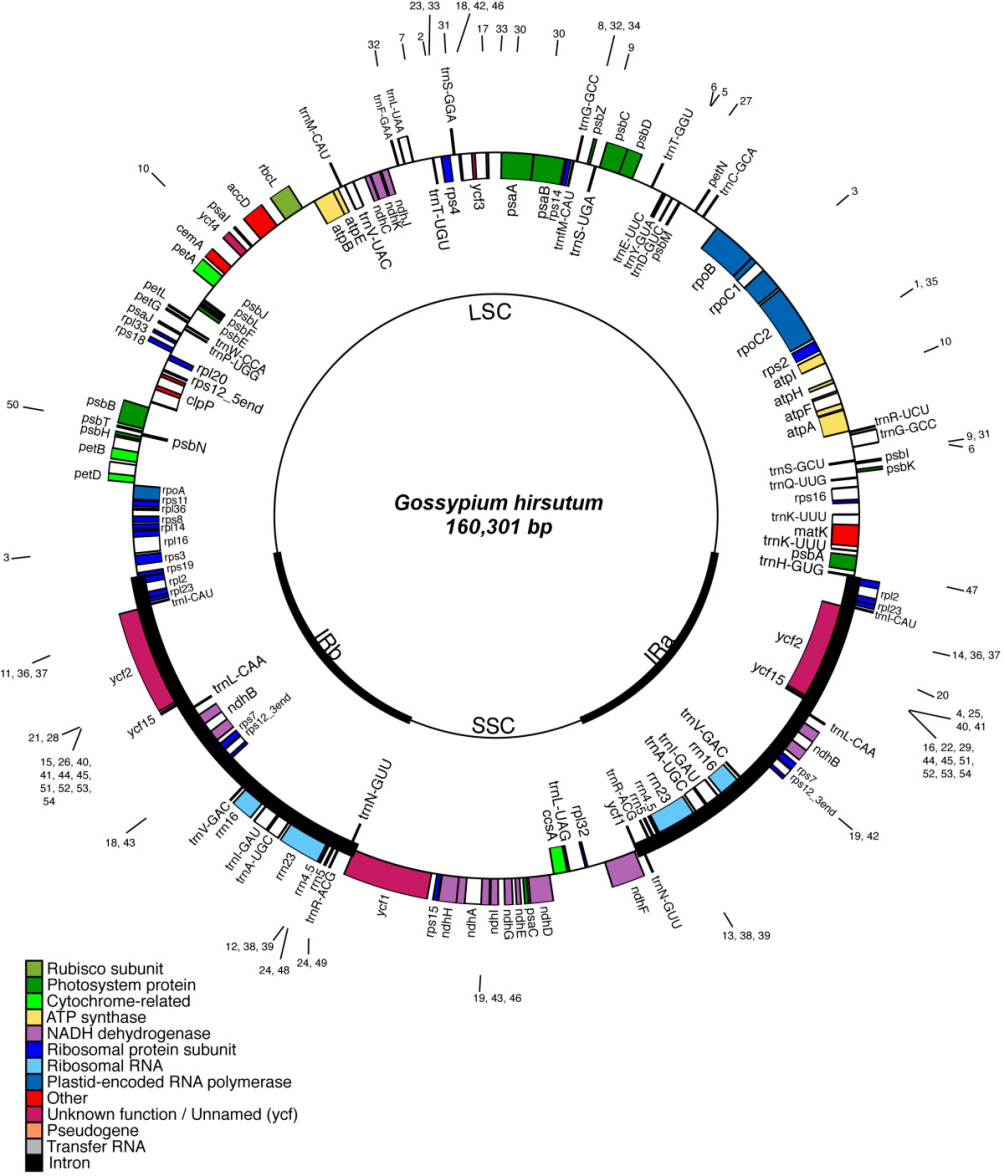
1. Chloroplast
Chloroplasts contain multiple copies of a circular genome and ribosomes that synthesize organelle-encoded proteins.
The chloroplast genome is circular and 120,000 to 160,000 bp depending on the species. No histones.
It includes ~120 genes:
- ~60 are involved in transcription and translation.
- About 20 genes are involved in photosynthesis.
- The RNA polymerases and ribosomal proteins are similar to those of bacteria.
Comparisons across plant species show variation whether some proteins are encoded in the chloroplast or the nucleus, but all chloroplasts contain similar sets of proteins. Therefore, some exchange of genes occurs between the chloroplast and nucleus.
2. Mitochondria
At mitosis, each daughter cell receives approximately the same number of mitochondria.
Each mitochondria contains multiple copies of the genome.
Therefore, the amount of mtDNA per cell depends on the number of mitochondria, the number of genomes per mitochondria, and the size of the mitochondrial genome.
To date, plant mitochondrial genomes have not been successfully transformed. Selection is a particular challenge because these organelles are essential for life.
遗传物质为DNA的病毒
Very simple “genomes” of just a few genes required for infection and replication. There are many, many types with different host specificities.
DNA viruses can also package their DNA , taking advantage of host cellular machinery such as histones.
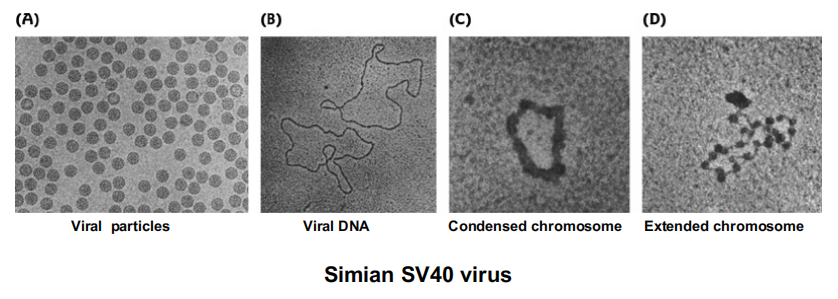
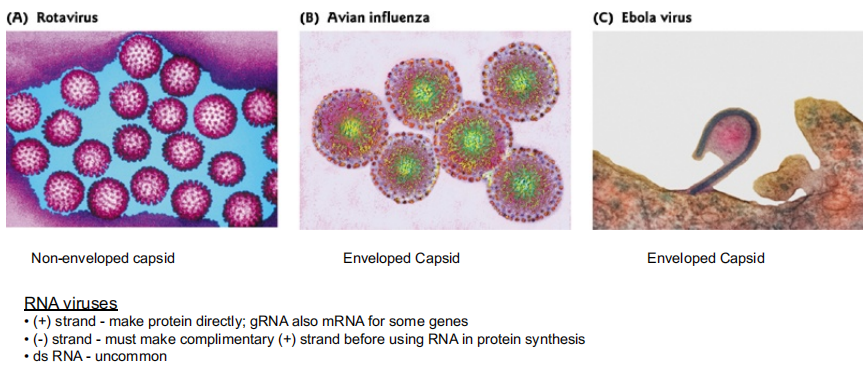
Eukaryotic RNA Viruses
核糖核酸病毒(英语:RNA virus),又称RNA病毒,其遗传物质为RNA,这些核糖核酸通常是单链RNA(ssRNA),但是也可能是双链RNA(dsRNA)。
Variety of sizes and shapes, enveloped or not. Typically replicate without an RNA intermediate (RNA RNA).