L13 Functional analysis of genes
一、Understanding the expression of a gene with spatial and temporal resolution
Why It Matters ? Gene Expression
The central dogma – gene expression is one of the major contributing factor of protein expression, which finally makes a cell “the living cell”.
What causes disease ? What are the basis of cell differentiation ? What determines the quality of the rice that you eat every day ?
These questions almost inevitably links to the gene expression pattern and the underlying regulation mechanisms.
What are the common ways to investigate gene expression ?
- Northern Blot
- Reverse-transcription quantitative PCR (RT-qPCR)
- Gene-CHIP
- Next generation sequencing
- Third generation sequencing
How to obtain gene expression information with spatial and temporal resolution ?
- Reporter Gene fusion
- In-situ hybridization
- Immunostaining
- Single Cell sequencing
Reporter Gene fusion
1. Reporter Gene fusion: GFP-target gene fusion
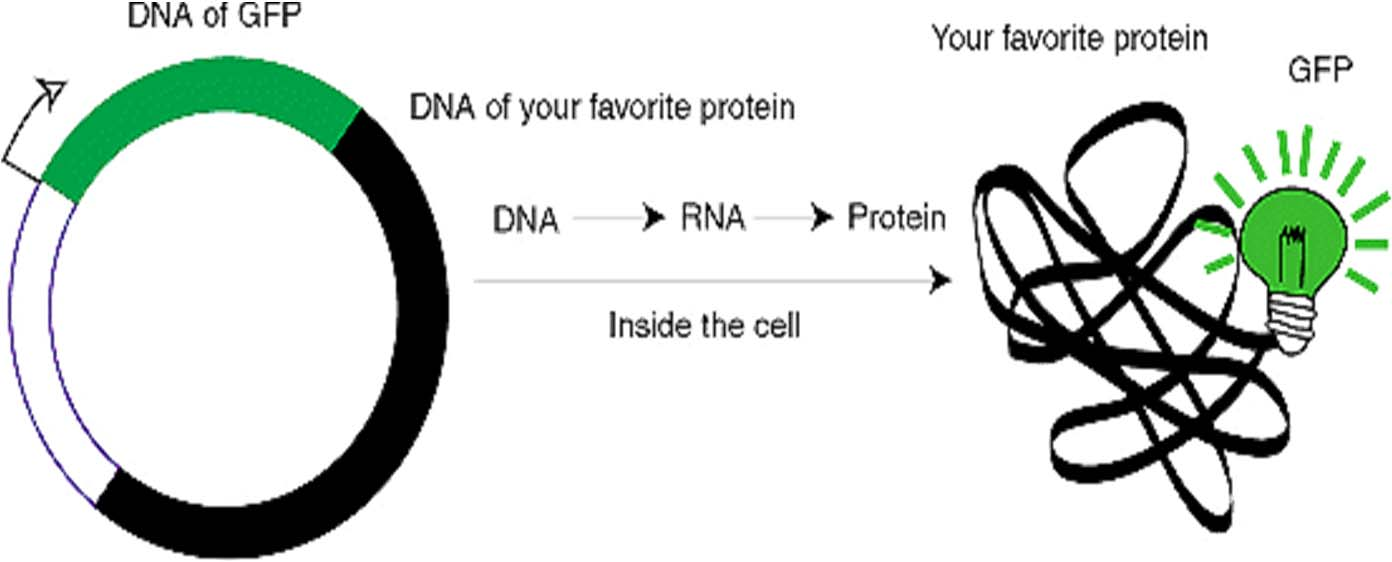
- Why make a GFP fusion ?
To track and quantify proteins (with subcellular resolution)
To watch protein-protein interactions
To describe biological events and signals in a cell

- GFP Was discovered by Osamu Shimomura in 1961
- GFP gene was cloned in 1992 by Douglas Prasher. In 1995, by changing a single amino acid, Roger Tsien engineered the first improved mutant of GFP with increased fluorescence and photostability、
- Today, many more derivatives have been created from GFP and dsRed (a red fluorescent protein)
2. Histochemical GUS assay
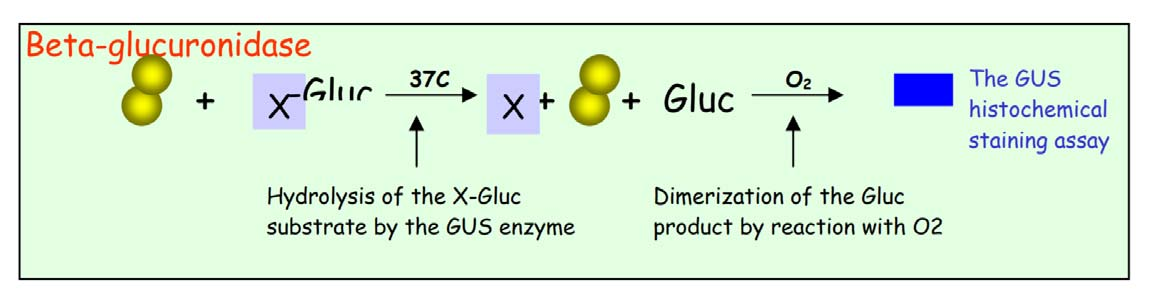
- GUS is short for β-Glucuronidase, a gene from Escherichia coli (E.coli) that encodes a sugar consuming enzyme–Glucuronidase. As there is no detectable GUS gene expression in plants, and the bacterial GUS gene does not interfere with plant intrinsic gene expressions, GUS provides an ideal reporter, being orthogonal to native genetic machinery, for gene expression in the plant system. By fusing the promoter of genes of interest to the GUS coding region, the expression patterns of plant genes can be reflected by the activation of GUS
X-Gluc (5-bromo-4-chloro-3-indolyl glucuronide) —–Beta-glucuronidase
X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) —–Beta-galactosidase
In-situ hybridization
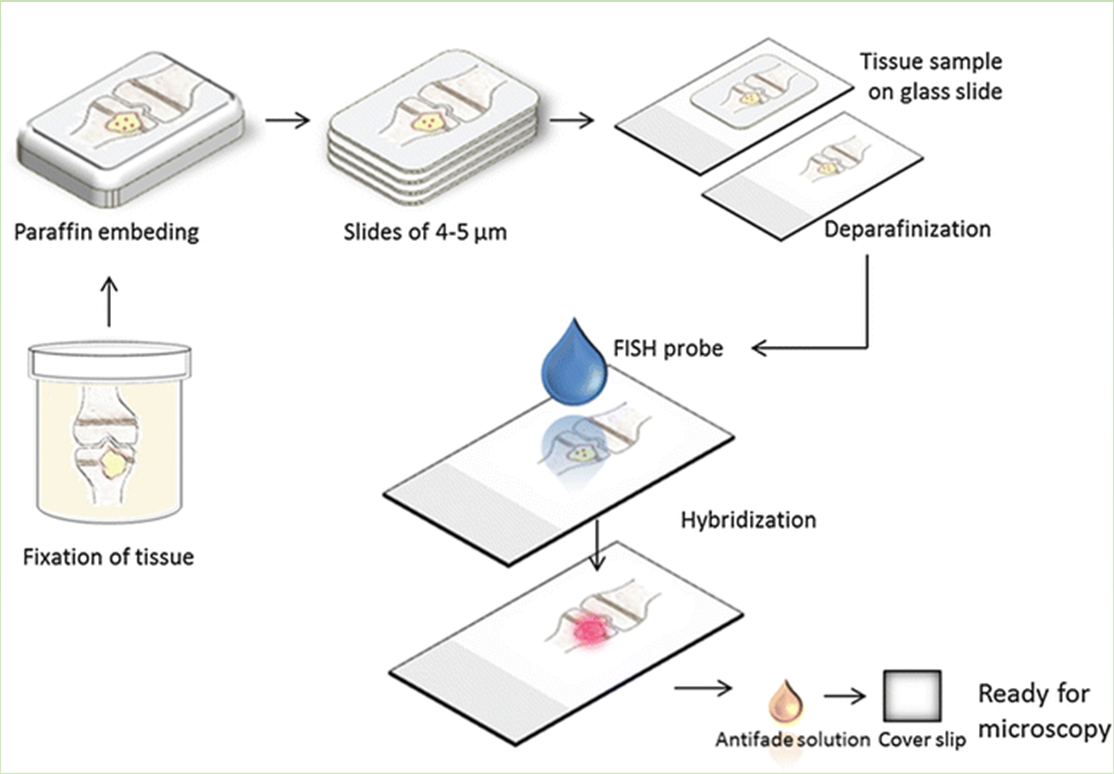
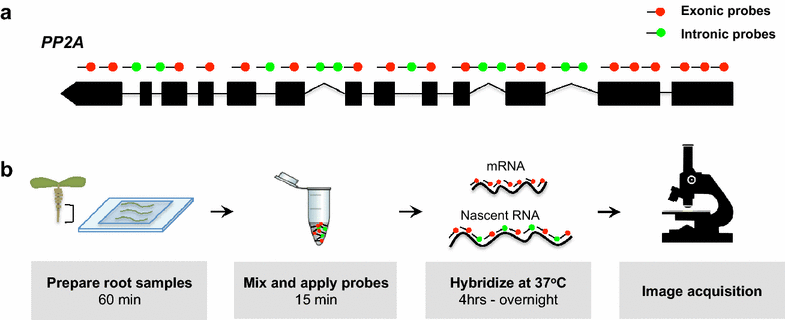
In situ hybridization enables the detection and precise localization of a specific nucleic acid sequence within an individual cell.
The nucleic acid sequence is bound specifically in a tissue section by complementary base pairing, that is, hybridization, with a detectable nucleic acid segment called a probe
In situ hybridization (ISH) combines three main advantages: great sensitivity, precise anatomical localization, and the possibility of quantification.
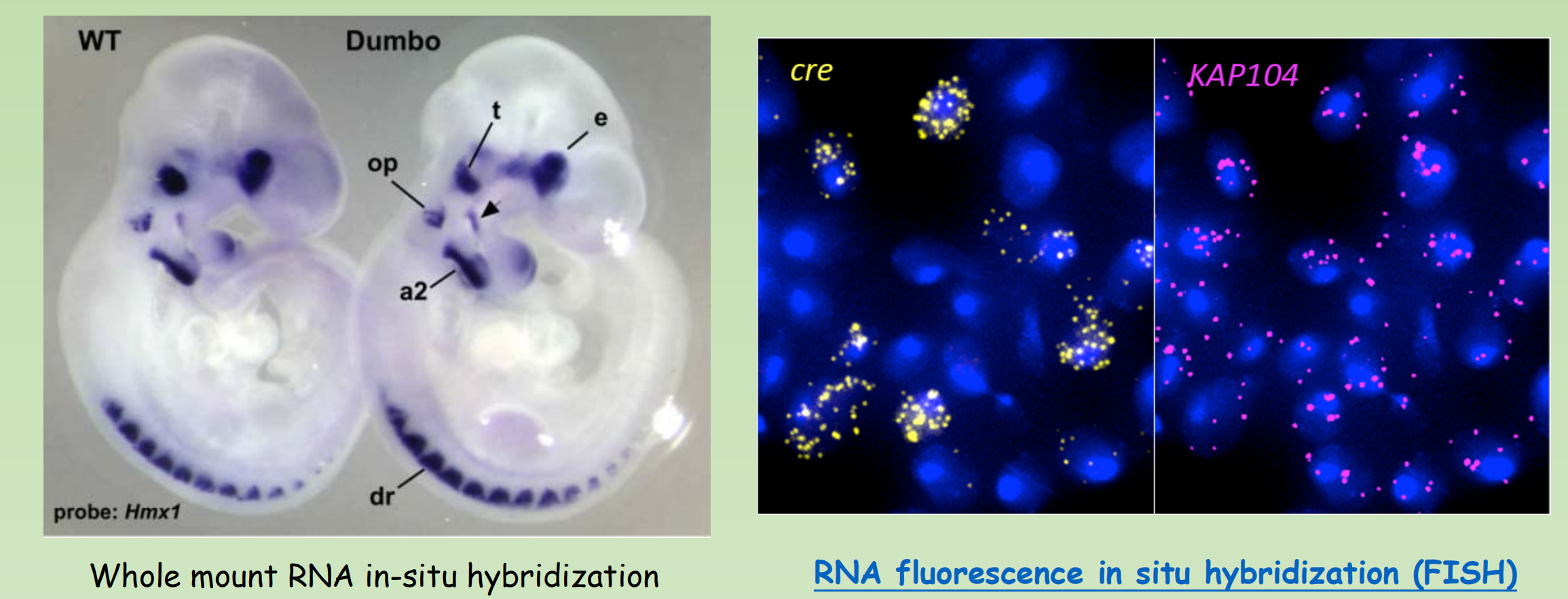
Single-molecule fluorescence in situ hybridization (smFISH)
- smFISH was developed to maximize both sensitivity and specificity by using multiple singly labelled probes to visualize RNA molecules as discrete spots of fluorescence
- A recent version of this method uses 48 fluorescently labeled DNA oligonucleotides (20mers) to hybridize to different portions of each transcript
- Preserves the spatial information of the cellular mRNA content with subcellular resolution within tissues.
Immunostaining
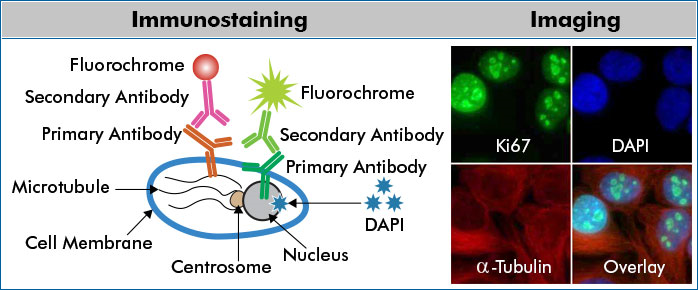
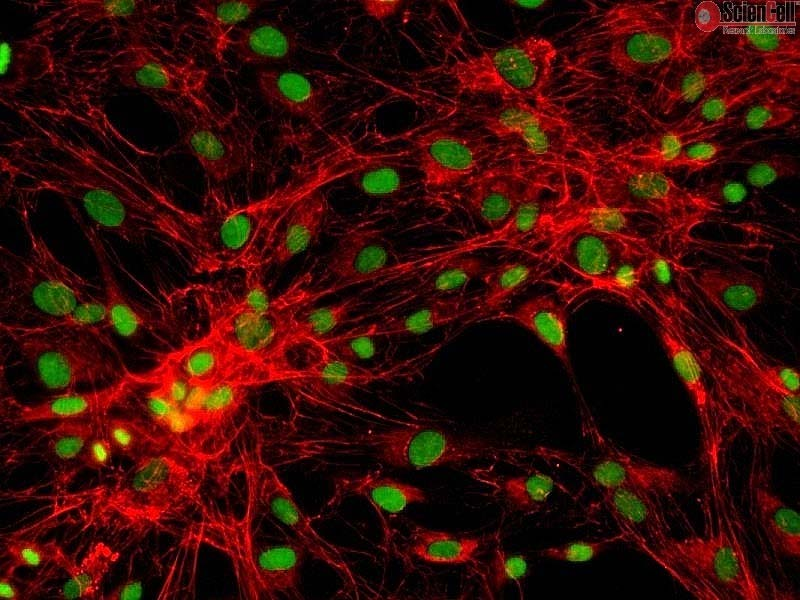
- Immunostaining uses antibodies to detect an antigen in cells or tissue.
- The major benefit of immunostaining in immunohistochemistry (IHC) is the ability to see the desired target in a tissue sample while maintaining the spatial context and tissue architecture.
Single Cell sequencing
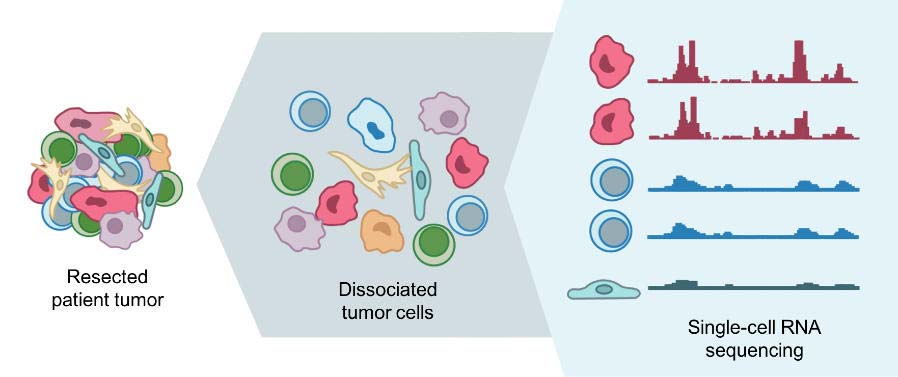
- Single cell sequencing examines the s equence information from individual c ells with optimized next-generation sequencing (NGS) technol ogies, providing a higher resolution of cellular differences and a better und erstanding of the function of an indiv idual cell in the context of its microe nvironment.
- In-situ RNA sequencing ?
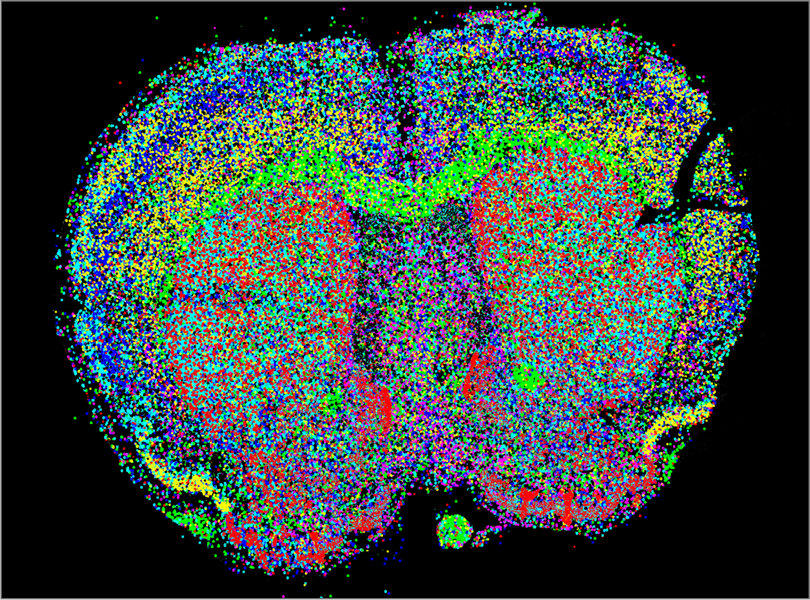
- In situ sequencing techniques detect individual gene transcripts (colored dots) in preserved mouse brain tissue. This approach allows researchers to identify and characterize different cell types in their native context with unprecedented detail. Xiaoyan Qian/Stockholm University
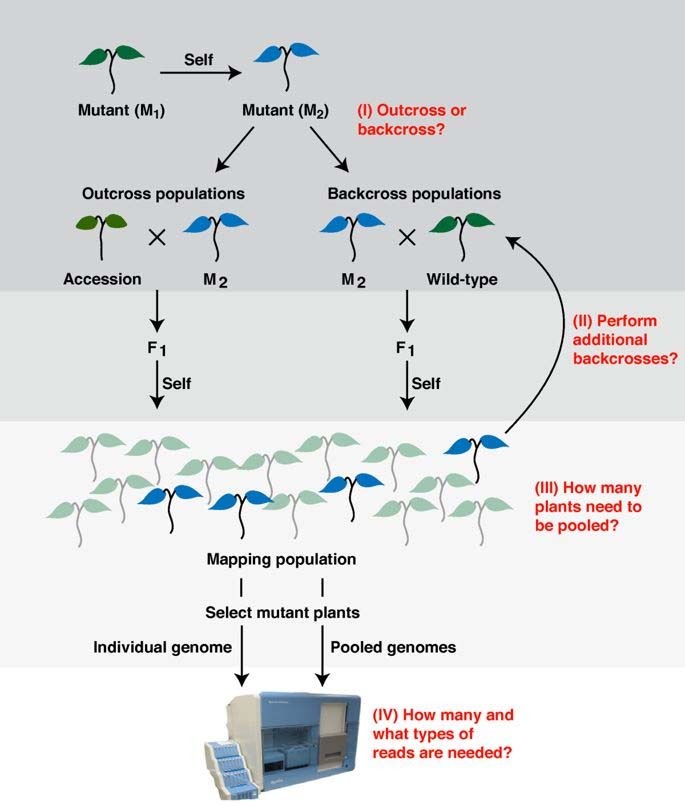
二、Build up a link between genotype and phenotype
Principle of Genetic mapping
How to identify the gene responsible for a certain phenotype ?
Genetic mapping is based on the use of genetic techniques to construct maps showing the positions of genes and other sequence features on a genome
Genetic techniques include cross-breeding experiments or, Case of humans, the examination of family histories (pedigrees).
Map-based cloning, also called positional cloning, is the process of identifying the genetic basis of a mutant phenotype by looking for linkage to markers whose physical location in the genome is known.
- Marker连接到多态性位点(Polymorphism),通过分析后代linkage的情况得知marker的相对位置
分析两个基因的linkage情况(WT和Mutant亲代纯合):
- 在F2代,PCR后跑胶的结果A、B基因会分别显示1:2:1的情况
- 若A与B连锁(linkage),那么出现A纯合(只有300bp)的情况下一定会伴随纯的400bp条带
Genetic Map
- A genetic map is a type of chromosome map that shows the relative locations of genes and other important features.
- The map is based on the idea of linkage.
- The distance between two markers on the genetic map indicate the probability that they will be inherited together, which means that the closer two genes are to each other on the chromosome, the greater the probability that they will be inherited together.
- By following inheritance patterns, the relative locations of genes along the chromosome are established
Physical Map
- A physical map of a chromosome or a genome that shows the physical locations of genes and other DNA sequences of interest.
- Physical maps are used to help scientists identify and isolate genes by positional cloning
Genetic Markers
- A genetic marker is a DNA sequence with a known physical location on a chromosome
- Genetic markers are used to track the inheritance of a nearby gene that has not yet been identified, but whose approximate location is known.
- The genetic marker itself may be a part of a gene or may have no known function
A example of genetic marker: Restriction fragment length polymorphisms (RFLP)
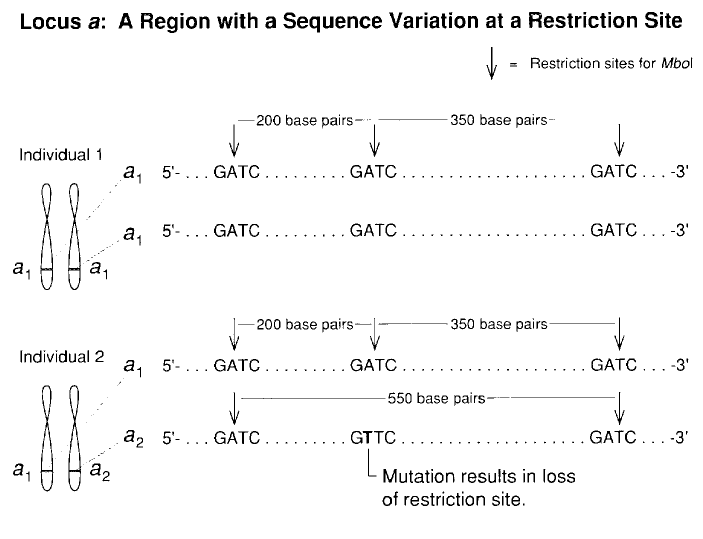
1. The procedure of map-based cloning
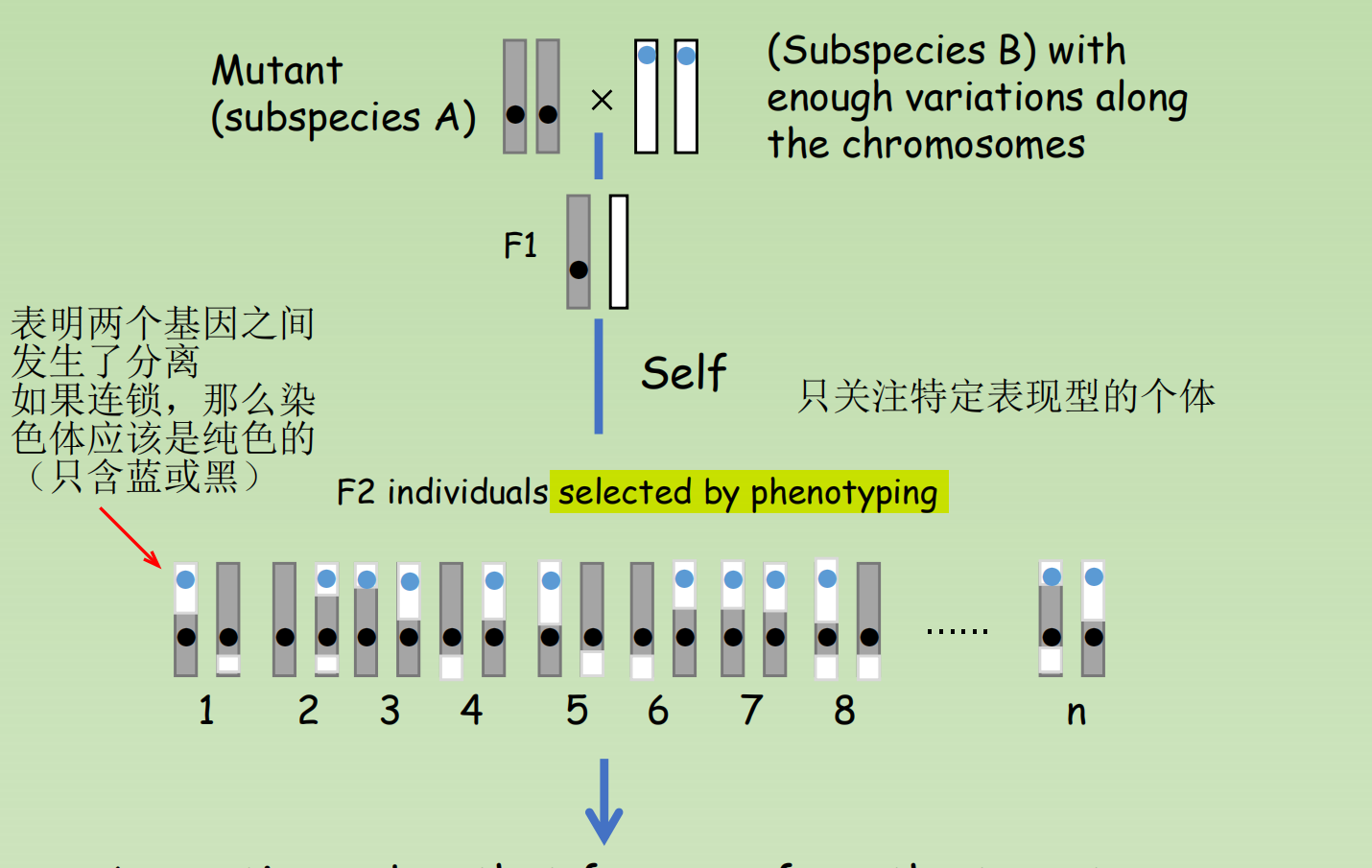
- A genetic marker that far away from the target gene would segregate according to the “law of segregation”
- A genetic marker that adjacent to the target gene would strictly inherent marker from subspecies A, with absence of the marker from subspecies B
- Therefore, we can sequence the region close to that genetic marker to identify the gene of our interest
Gene Knock-out and Gene Knock-in
After you cloned a gene through mapped based cloning, how can you be sure that you indeed isolated the gene that responsible for the phenotype ?
There are often two ways
- One can introduce a functional copy of the candidate gene and see if the phenotype has been complimented. this is known as complementation test
- The action that artificially introduce a (foreign) gene into the cell is known as Gene Knock-in
- Alternatively, by using a wild-type organism, one can try to deplete or inactivate the gene of interest, the phenotype should be reproduced through this way.
- The action that a gene is completed being inactivated or depleted is known as Gene knock-out
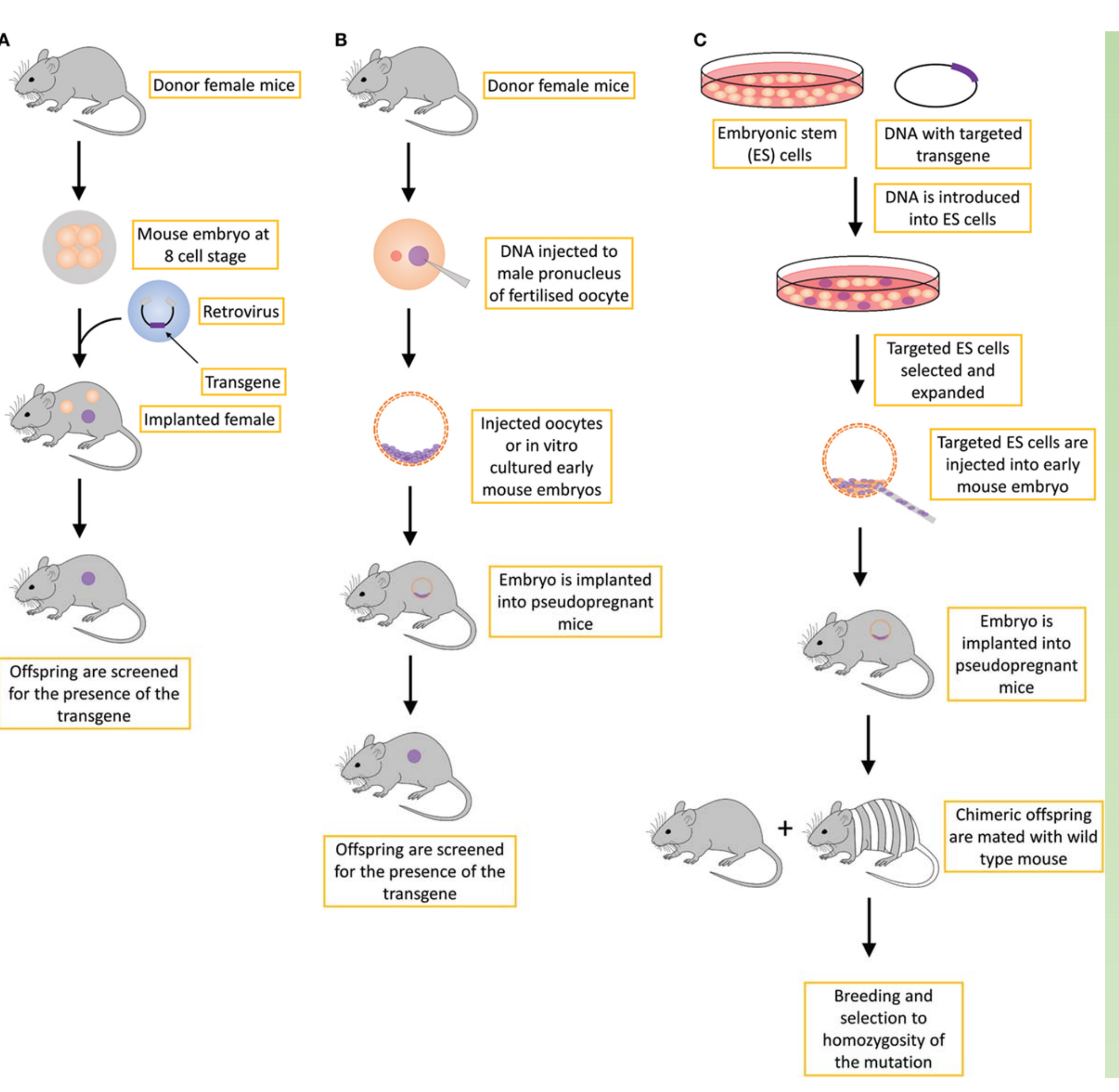
Different strategies for creating transgenic mice
- (A) retroviral approach, which is not routinely used;
- (B) standard transgene approach, in which the DNA is inserted into the genome in an unspecific manner;
- (C) gene-targeted transgene approach, which is an approach that is routinely used to create conventional knockout transgenic mice, usually with a constitutive loss-of-function mutation.
Gene Knock-out and Gene Knock-in has been absolutely important for modern biology

1. Principle of making a knock-out Mice
Which method is better ?
Gene Targeting - Adv. - if the DNA sequence of the target gene is known, researchers can precisely knock out the gene at a high rate of efficiency
Gene Trapping – Adv. researchers do not need to know the DNA sequences of specific genes in order to knock them out. Also, a single Vector can be used to knock out various genes
- 基因陷阱:随机敲击,后鉴定
Disadv. not every successful insertion of artificial DNA into a gene leads to a loss of function. Also considerable time must be spent conducting tests to identify ES cells in which gene(s) actually have been knocked out
- So Gene Targeting (by homologous recombination) is widely used.
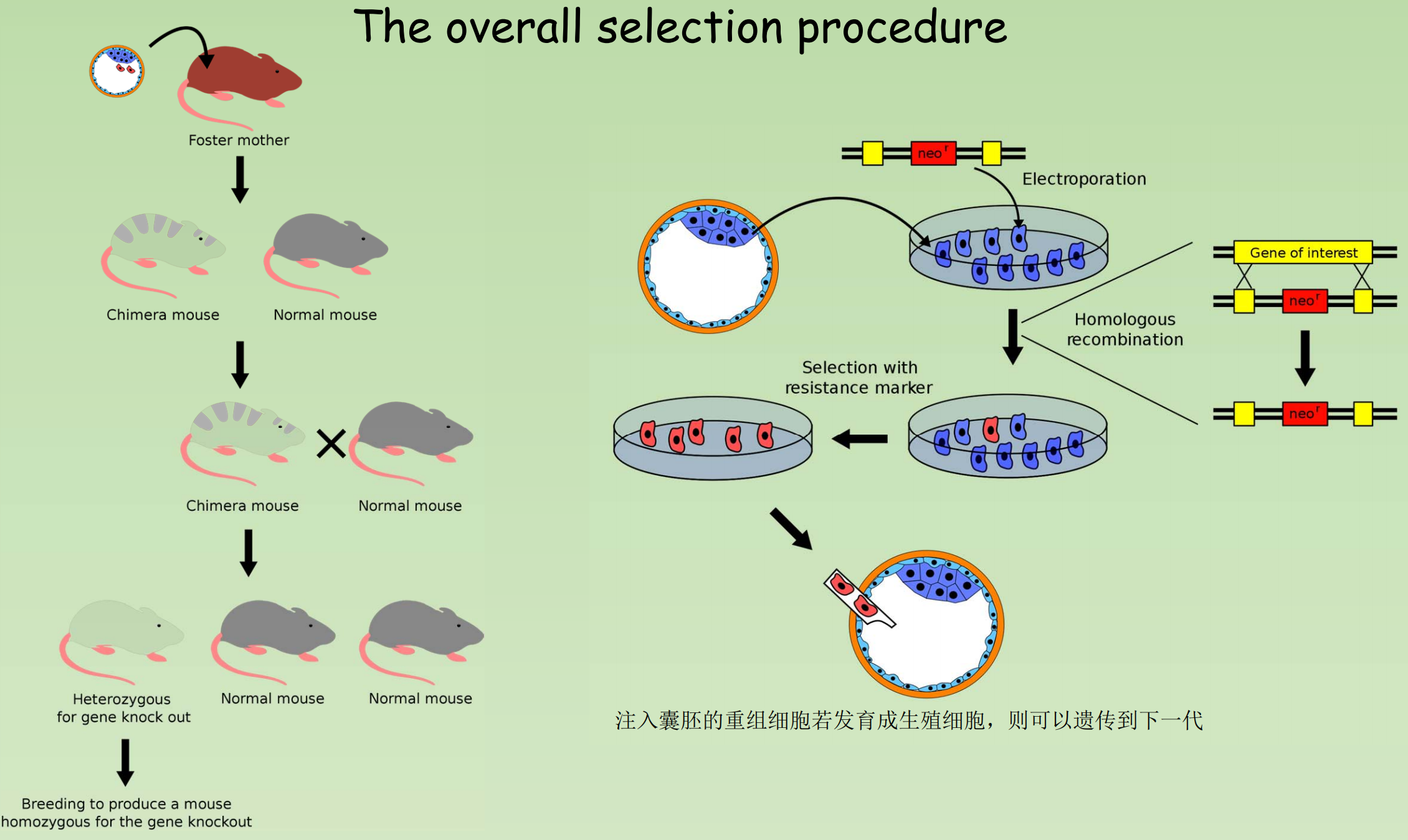
Production of knockout mice (by Gene Targeting)
- Harvesting of Embryonic stem cells from a mouse blastocyst.
- Introducing the artificial DNA into the ES cells in the culture.
- Screen ES cells and select those whose DNA includes the new gene
- Implant selected cells into normal mouse embryos, making “chimeras“
- Implant chimeric embryos in pseudopregnant females.
- Females give birth to chimeric offspring, which are subsequently bred to verify transmission of the new gene, producing a mutant mouse line.
(1) Positive selection
筛选出发生了重组的细胞
- It involves the isolation of a target cell population by using an antibody that specifically binds that population.
- Positive selection markers are used to enrich for recombination events
- Eg. Encoding Antibiotic resistance gene Neomycin
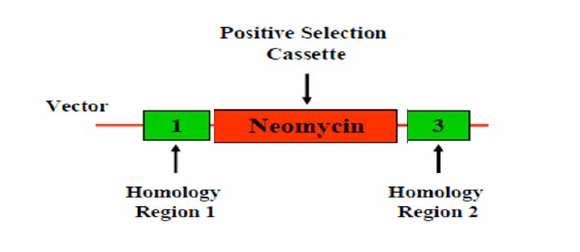
- Flanked by two arms of homologous sequence
- Enrich recombination events through Neomycin selection.
- Fragment outside of homologous arm won’t be inserted into host genome.
(2) Negative selection
筛选出没有发生变化的细胞
- It involves the depletion of all cell types except your cell type of interest
- Negative selection markers used to enrich for homologous recombination events over random insertions
- Eg. Use of Herpes Simplex Virus (HSV) Thymidine Kinase (TK) gene coupled with gancyclovir treatment
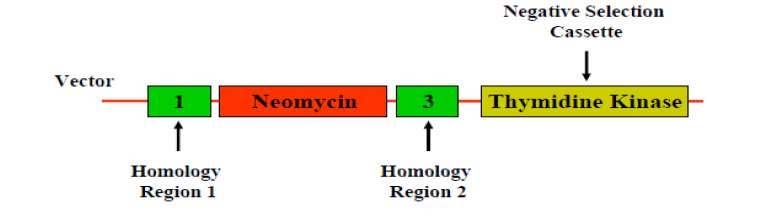
- In case the fragment from vector insert into genome randomly.
- Negative selection kill these cells
- Thymidine Kinase sit outside the homologous arm
- The presence of Thymidine Kinase (TK, from Herpes Simplex Virus) gene coupled with ganciclovir treatment kills the cell.
(3) The overall selection procedure
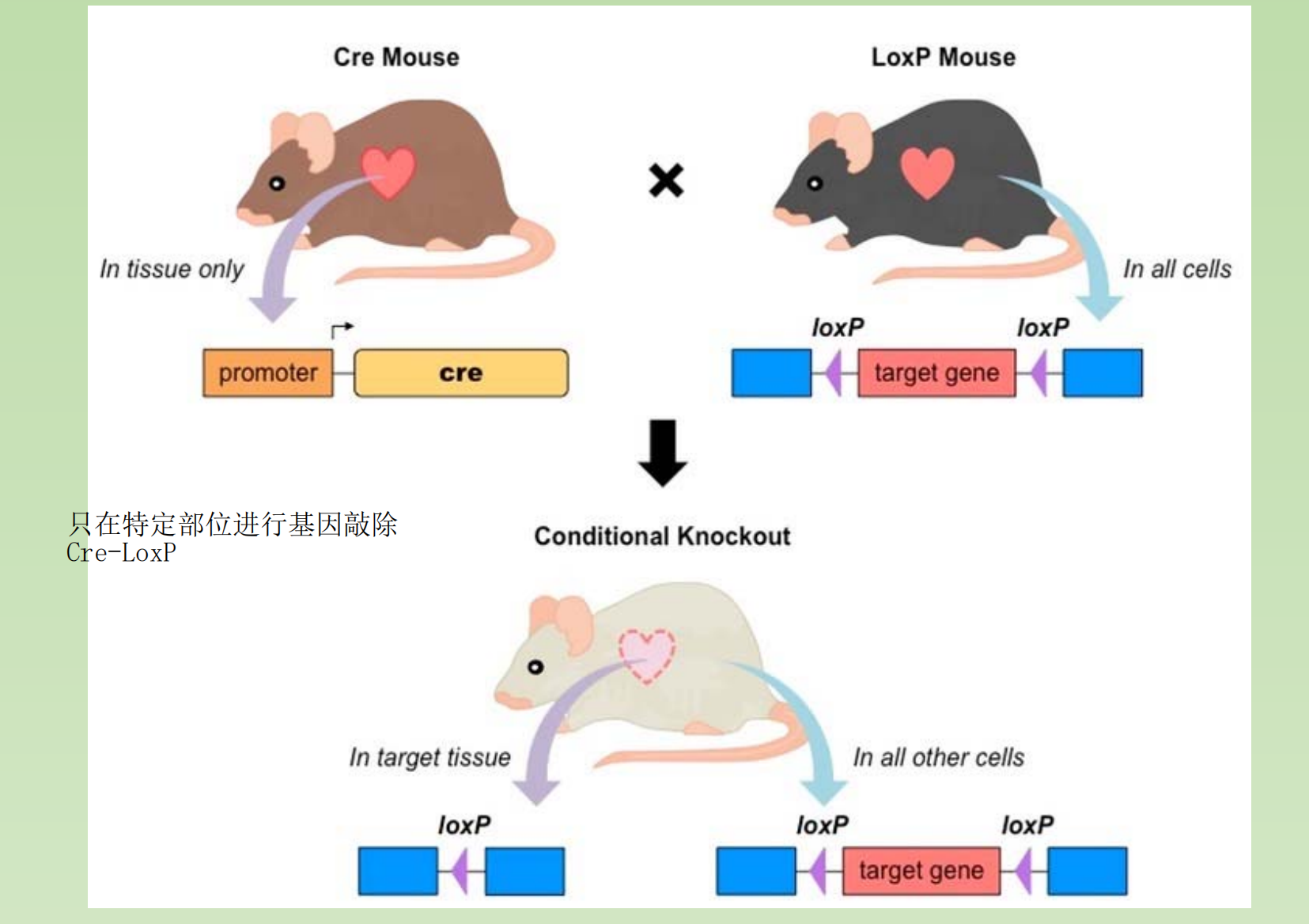
2. Conditional Knockout Mouse – A more accurate choice of gene knockout

GDF8 (Myostatin) knockout mouse:
- More than twice the muscle mass of a wildtype mouse
FGF5 knockout mouse:
- It has long, angora-like hair
Gene Knock-out and Gene Knock-in in Plant
Unlike the situation in mammalian cells, homologous recombination has poor efficiency in plant cell. There are two ways to introduce a foreign gene into plant cell.
- Mechanical force mediated transformation
- Agrobacterium tumefaciens mediated transformation
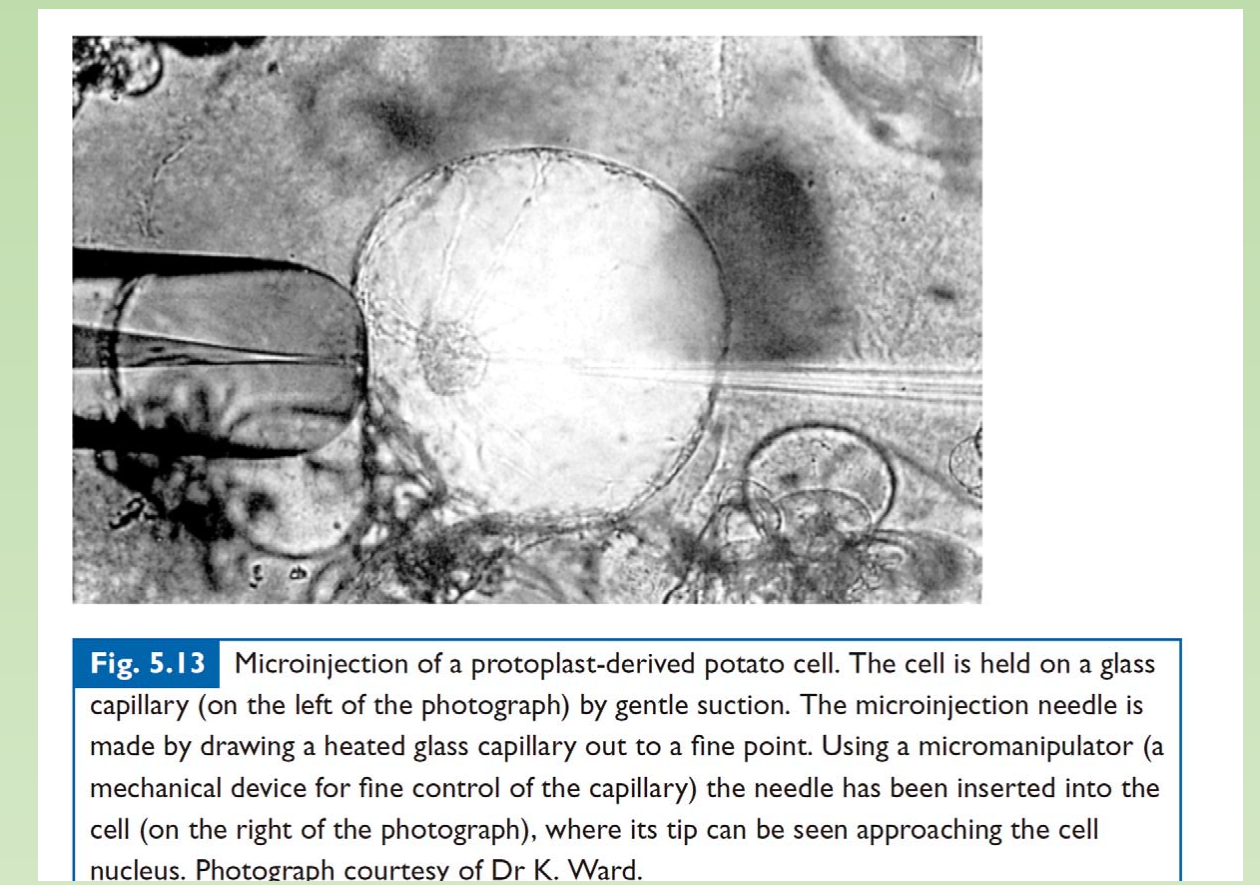
1. Microinjection
Biolistic Particle Delivery: Gene Guns or Biolistic Bombardment
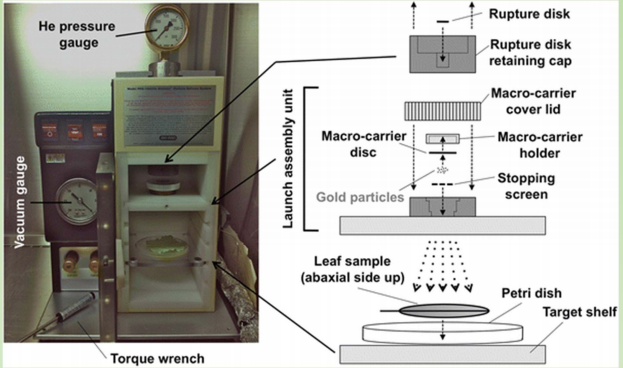
- Particles (e.g. gold) are coated with DNA (vector) and accelerated to a very high speed, and therefore the DNA is able to go into the host cell when the particle penetrate the plant tissue.
Gene Knock-out and Gene Knock-in in Plant
- Agrobacterium tumefaciens – pathogen and useful tool
Crown gall disease and the tumor-inducing principle
- 1907: Crown gall is caused by a bacterium
- Agrobacterium-induced galls do not require bacterial persistence
Gall tissues can grow indefinitely without exogenous phytohormones
2. Structure and function analysis of the Ti plasmid
Some DNA from the Ti plasmid is transferred into the plant cells (1977)
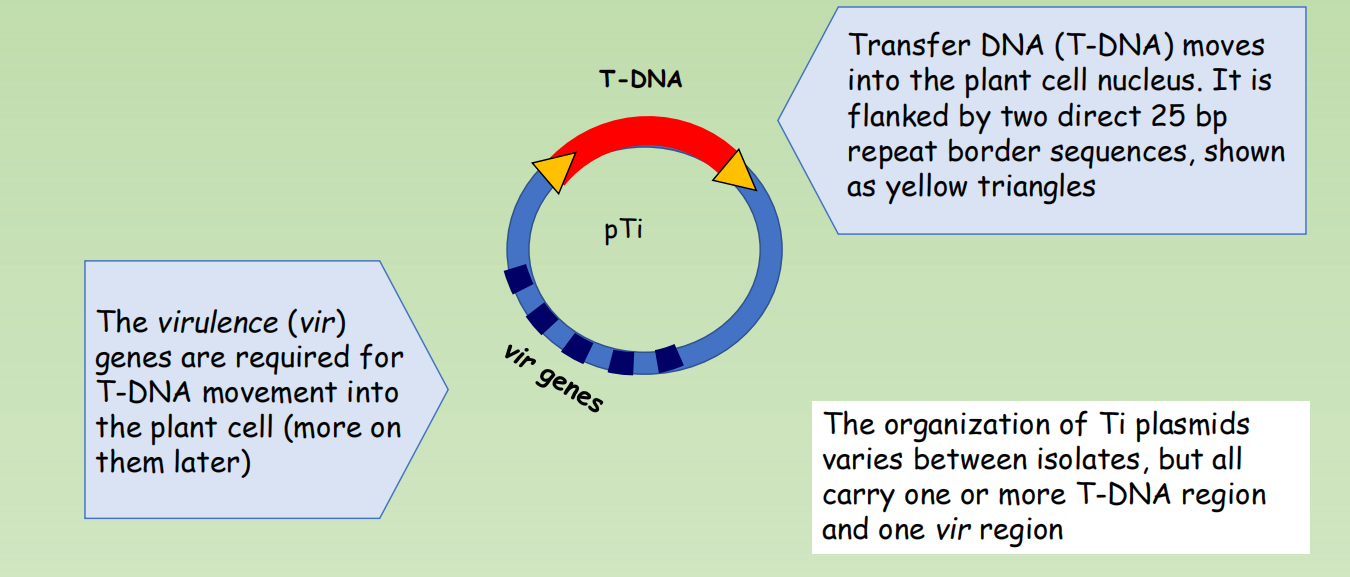
- Transfer DNA (T-DNA) moves into the plant cell nucleus. It is flanked by two direct 25 bp repeat border sequences, shown as yellow triangles
- The virulence (vir) genes are required for T-DNA movement into the plant cell (more on them later)
- The organization of Ti plasmids varies between isolates, but all carry one or more T-DNA region and one vir region
The T-DNA region: tumor-inducing genes and opine synthesis genes
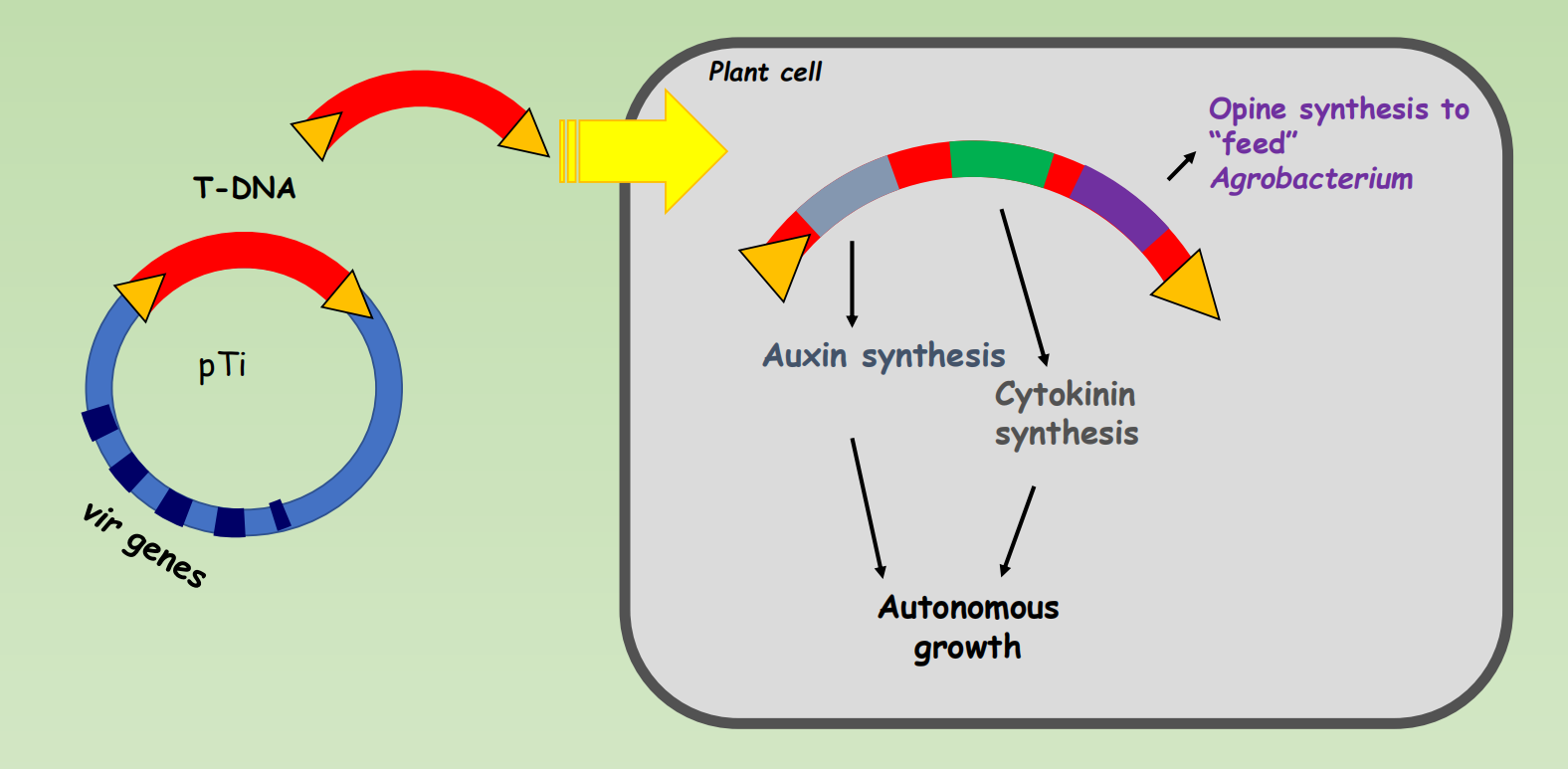
The Ti plasmid can be used to introduce any gene into plants
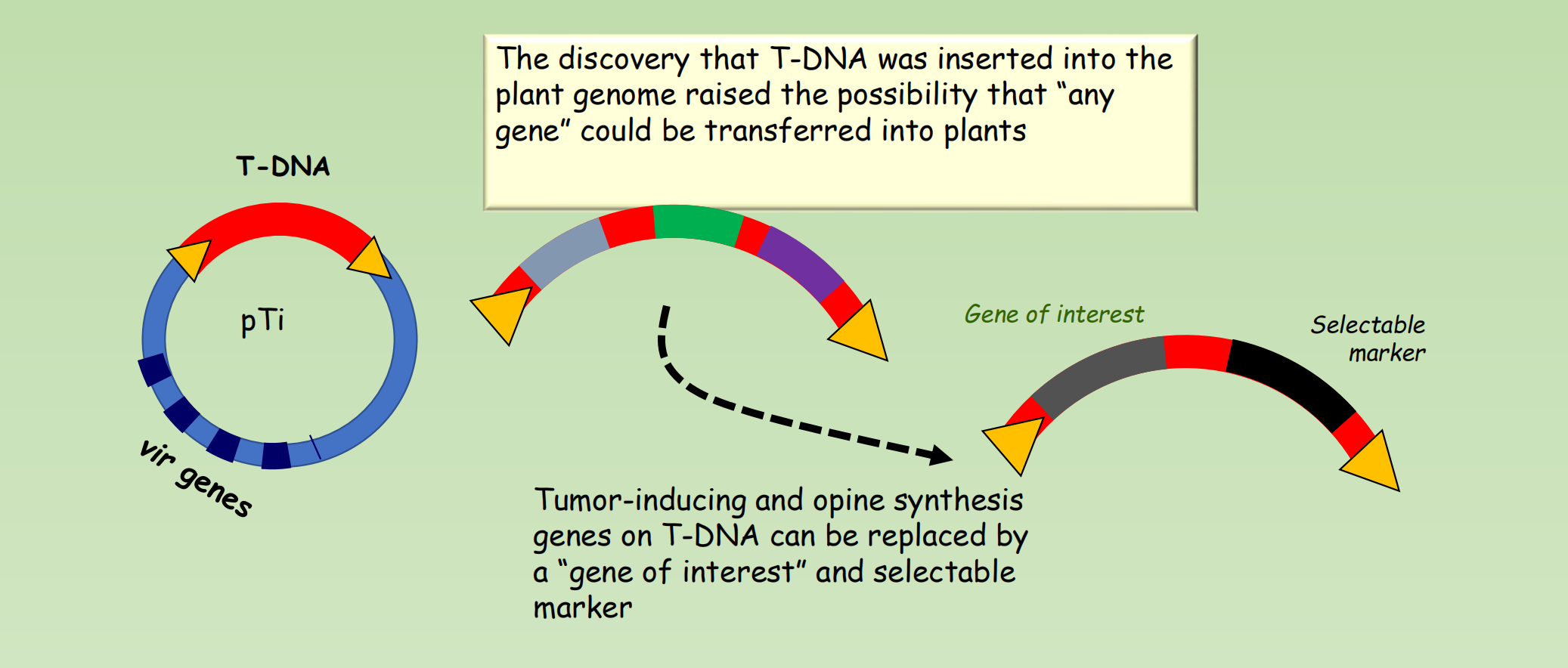
- The discovery that T-DNA was inserted into the plant genome raised the possibility that “any gene” could be transferred into plants
- Tumor-inducing and opine synthesis genes on T-DNA can be replaced by a “gene of interest” and selectable marker
Development of binary vectors that separate T-DNA and vir genes
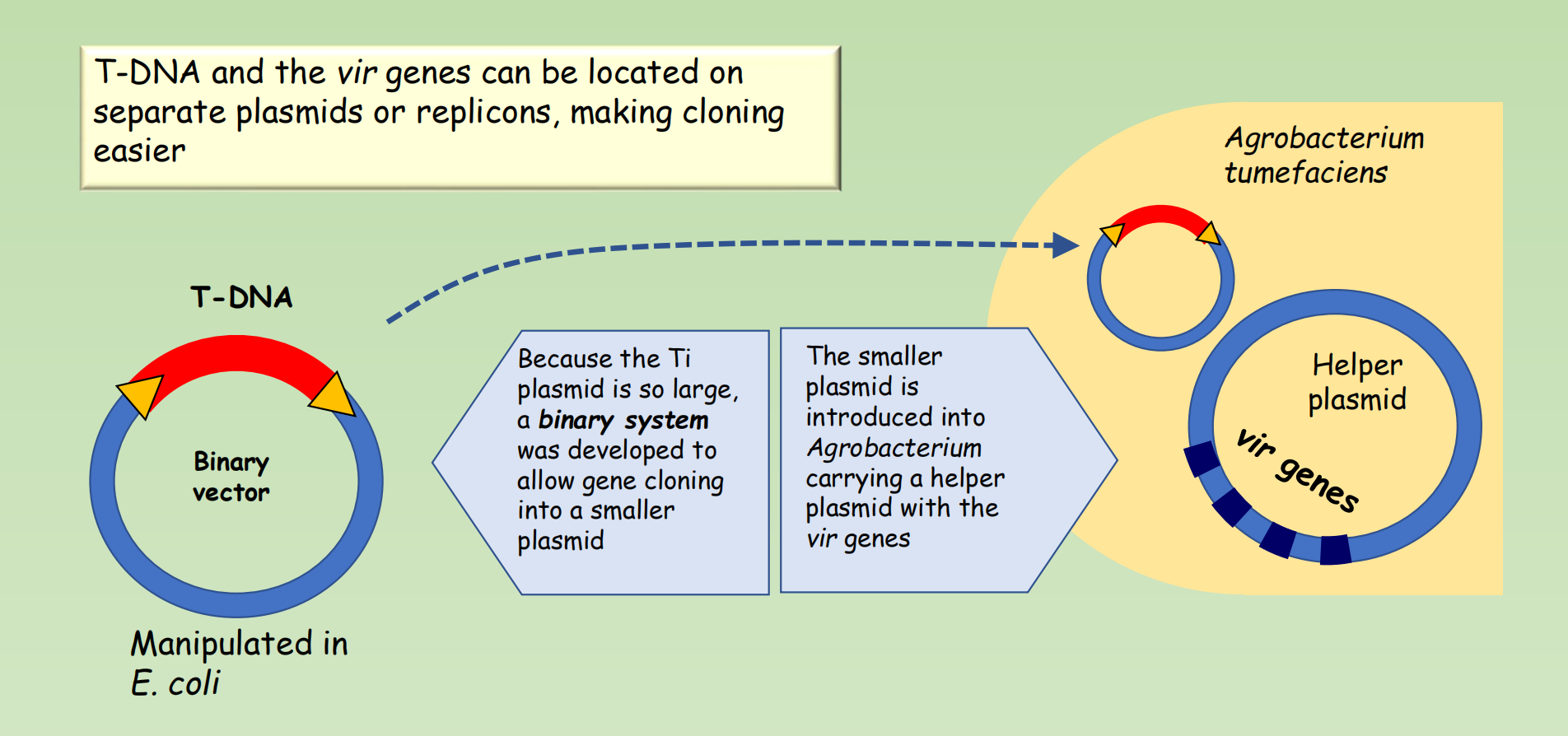
- T-DNA and the vir genes can be located on separate plasmids or replicons, making cloning easier
- Because the Ti plasmid is so large, a binary system was developed to allow gene cloning into a smaller plasmid
- The smaller plasmid is introduced into Agrobacterium carrying a helper plasmid with the vir genes
3. Regeneration of transgenic plants from transformed cells
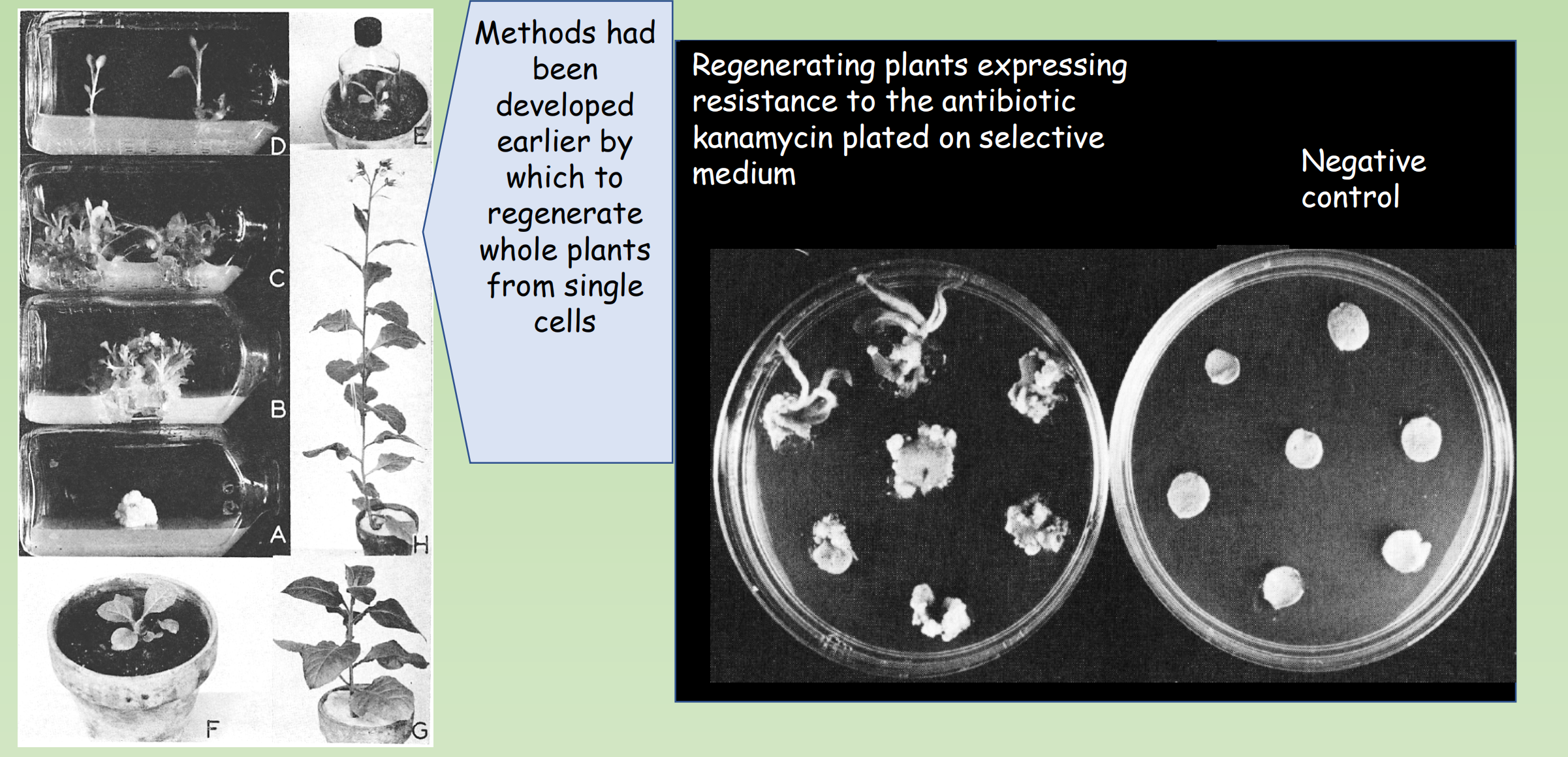
- Methods had been developed earlier by which to regenerate whole plants from single cells
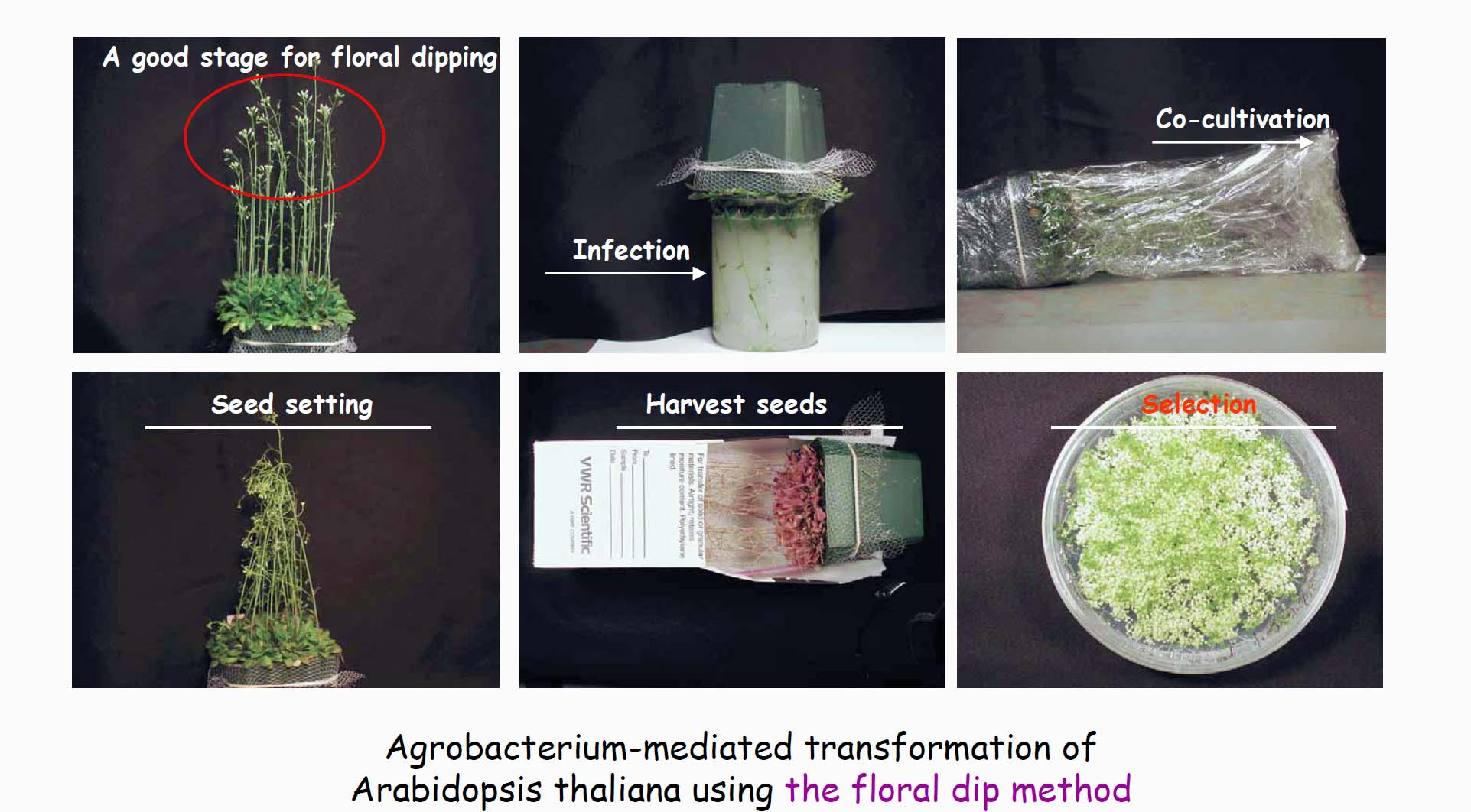
4. Agrobacterium-mediated transformation has other uses
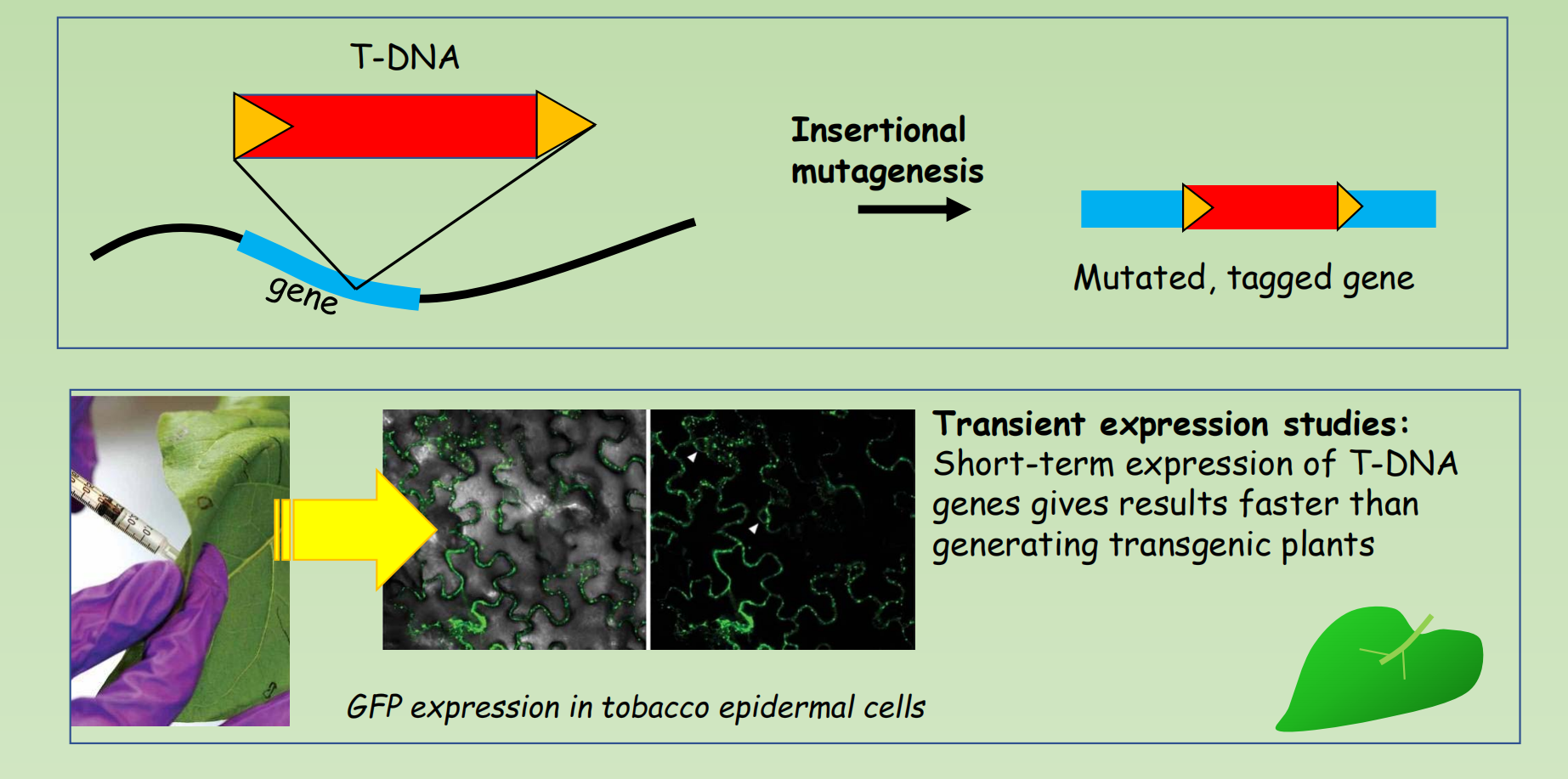
Transient expression studies:
- Short-term expression of T-DNA genes gives results faster than generating transgenic plants
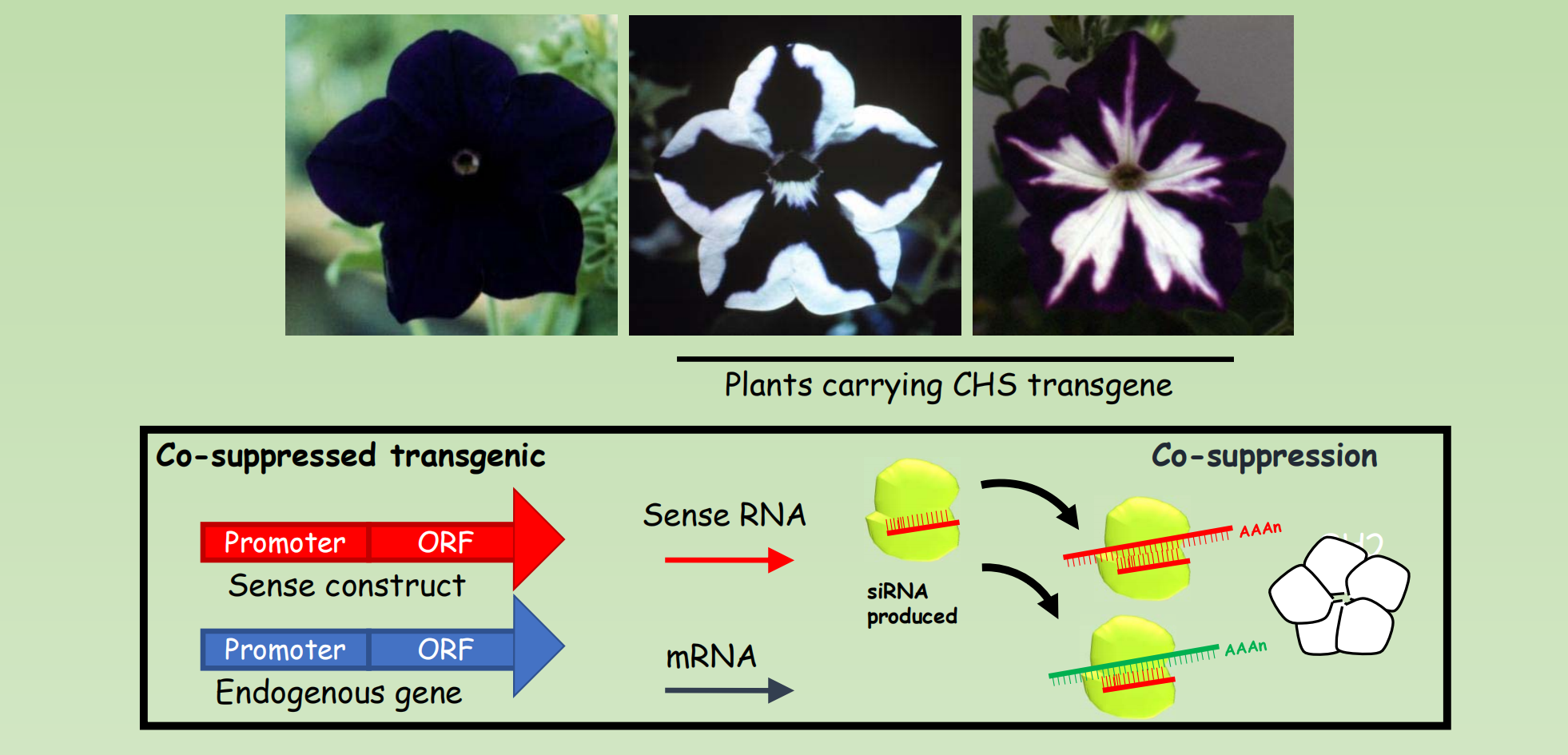
Random insertion of T-DNA create a chance for the isolation of Knock-out mutant of one third of genes in Arabidopsis

Epigenetic gene silencing was discovered in transformed plants