L10 Intracellular transport II - ER and Golgi
一、ER import
Structure of ER
All eukaryotic cells have an endoplasmic reticulum (ER). Its membrane typically constitutes more than half of the total membrane of an average animal cell
Thus, the ER and nuclear membranes form a continuous sheet enclosing a single internal space, called the ER lumen or the ER cisternal space, which often occupies more than 10% of the total cell volume
The ER membrane is also the site at which most of the lipids for mitochondrial and peroxisomal membranes are made.
The ER Is Structurally and Functionally Diverse
1. Endoplasmic reticulum under light microscope
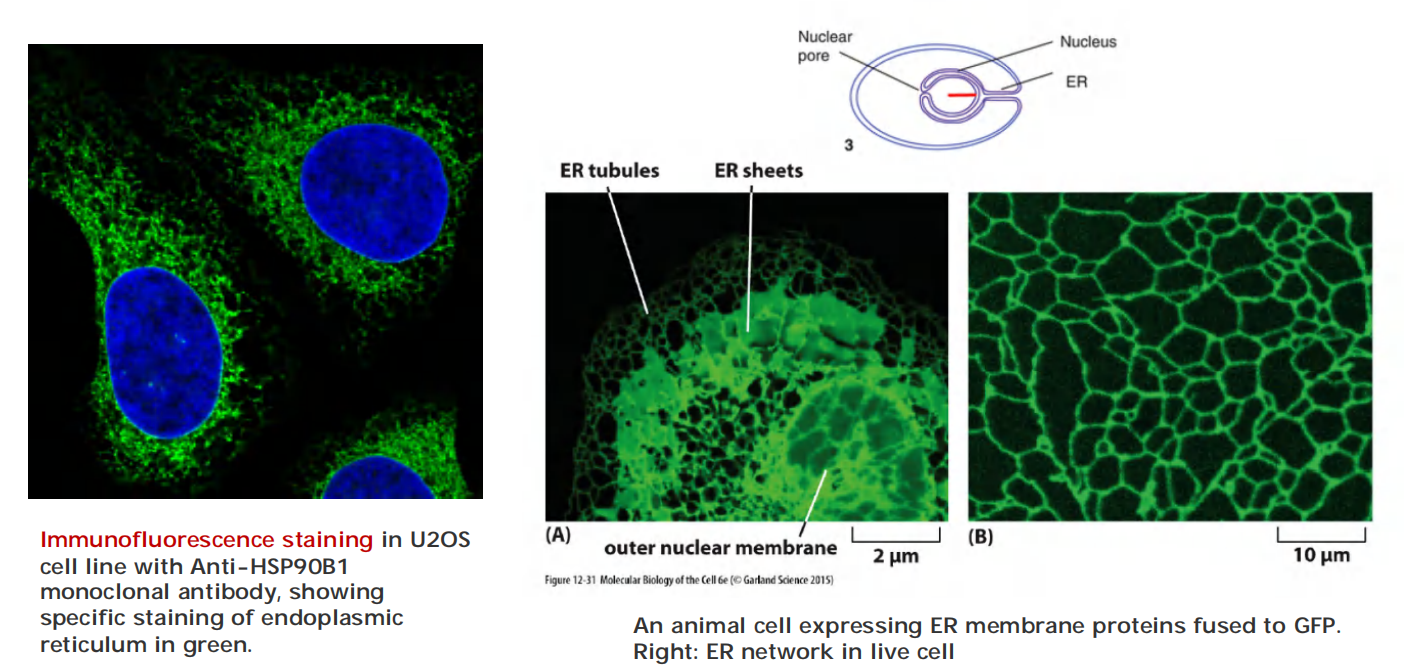
2. Endoplasmic reticulum under electron microscope
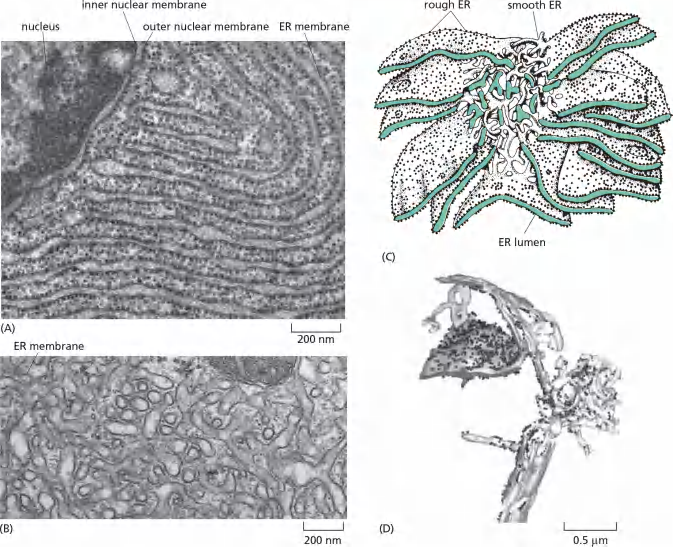
The ER possesses different regions/domains:
- smooth ER and rough ER are functionally and structurally different
- Some cell have partial rough and partial smooth ER
- Some cells have a scanty (稀疏的,少量的) region of smooth ER
- Some cells have abundant smooth ER (e. g., muscles cells)
- Some cells have expanded smooth ER to meet the function (synthesis of steroid hormone from cholesterol)
- Transitional ER (lecture 11)
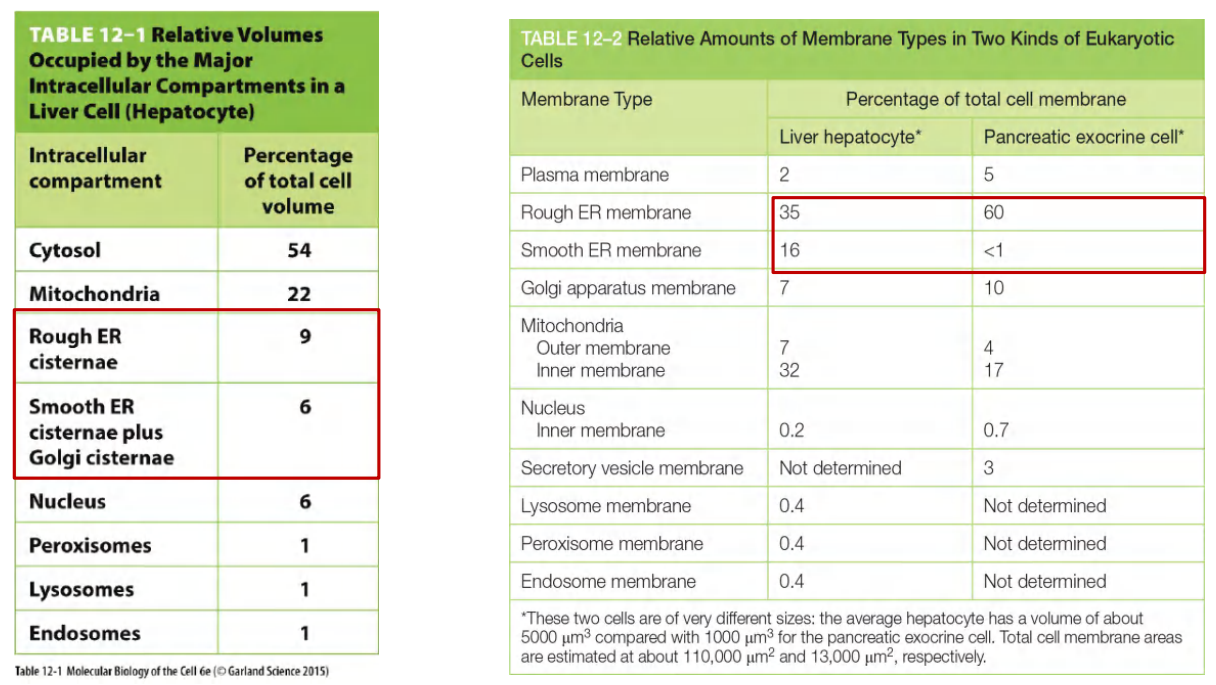
3. Volume in Different Cells
Endoplasmic reticulum (ER) occupies large volume and accounts for large amounts of the cell
Questions:
The ER possesses different regions/domains:
- smooth ER and rough ER are functionally and structurally different
- Why do ribosomes attach to ER but not to mitochondria?
- How proteins are imported into ER?
These membrane-bound ribosomes coat the surface of the ER, creating regions termed rough endoplasmic reticulum, or rough ER;
Regions of ER that lack bound ribosomes are called smooth endoplasmic reticulum, or smooth ER
ER Functions
To meet different functional demands, distinct regions of the ER become highly specialized
We observe such functional specialization as dramatic changes in ER structure, and different cell types can therefore possess characteristically different types of ER membrane. One of the most remarkable ER specializations is the rough ER
1. Featured Functions
Lipid synthesis: nearly all of the cell’s major classes (phospholipid, cholesterol, glycolipid precursor) of lipids are synthesized in ER.
Transmembrane proteins synthesis: Produce transmembrane proteins for ER, Golgi, endosome, lysosome, and plasma membrane
Lysosomes contain digestive enzymes that degrade defunct intracellular organelles, as well as macromolecules and particles taken in from outside the cell by endocytosis.
Water soluble proteins in the lumen of ER, endosome, and lysosome, respectively.
Secreted proteins synthesis
Store Ca2+ (Ca2+ pump and Ca2+ release channel, control intracellular Ca2+ concentration) (see lecture 7 and 8)
Another crucially important function of the ER in most eukaryotic cells is to sequester(使隔绝;使隔离) Ca2+ from the cytosol. The release of Ca2+ into the cytosol from the ER, and its subsequent reuptake, occurs in many rapid responses to extracellular signals
Muscle cells have an abundant, modified smooth ER called the sarcoplasmic reticulum (SR). The release and reuptake of Ca2+ by the sarcoplasmic reticulum trigger myofibril contraction and relaxation, respectively, during each round of muscle contraction
Detoxification: e.g. cytochrome p450
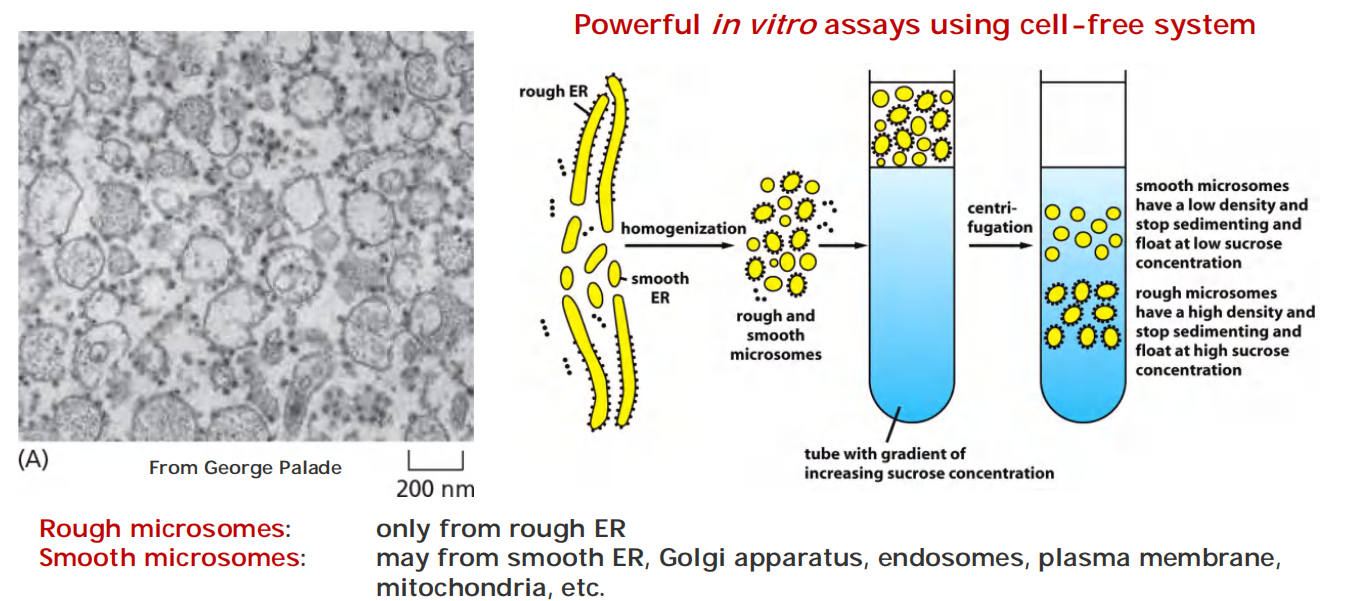
2. Isolation and purification of microsomes from ER
Microsomes (微粒体): physiologically intact
- capable of protein translocation, protein glycosylation, uptake of Ca2+ and release, lipid synthesis, etc.
Because of the unusually large quantities of smooth ER or sarcoplasmic reticulum, respectively, most of the smooth microsomes in homogenates(均匀混合物,(组织)匀浆) of these tissues are derived from the smooth ER or sarcoplasmic reticulum.
The signal hypothesis and discovery of signal peptides
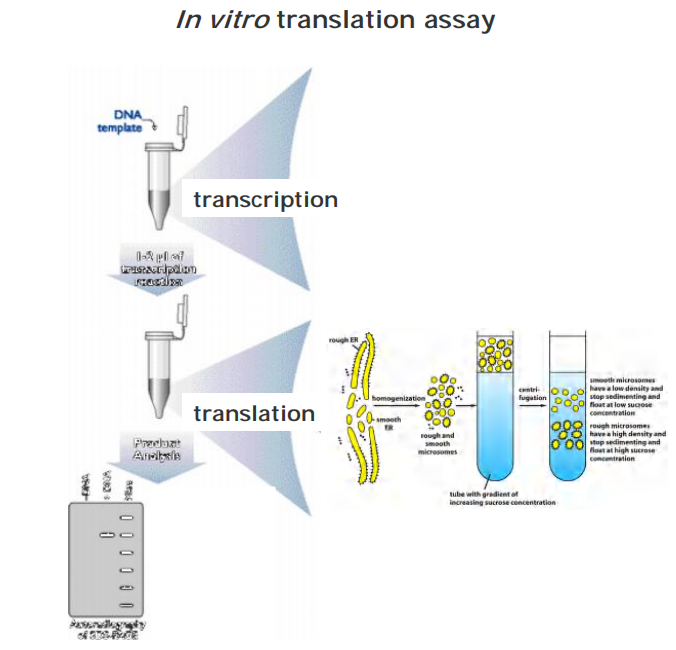
1970, study secreted proteins
In vitro translation assay (cell-free)
- mRNA of the secreted protein
- Ribosome
- With or without microsome
- Examine the translated protein by SDS-PAGE
Result: The size of the protein was smaller in the presence of microsome than that in the absence of microsome
- In the absence of microsomes, the protein synthesized was slightly larger than the normal secreted protein.
- In the presence of microsomes, the protein of correct size was produced.
These size difference suggests the presence of an initial signal sequence that guide the import of protein to the ER.
Signal peptides were first discovered in protein imported in rough ER proteins.
Route of ER Transport
Mammalian cells begin to import most proteins into the ER before complete synthesis of the polypeptide chain—that is, import is a co-translational process
- In contrast, the import of proteins into mitochondria, chloroplasts, nuclei, and peroxisomes is a post-translational process
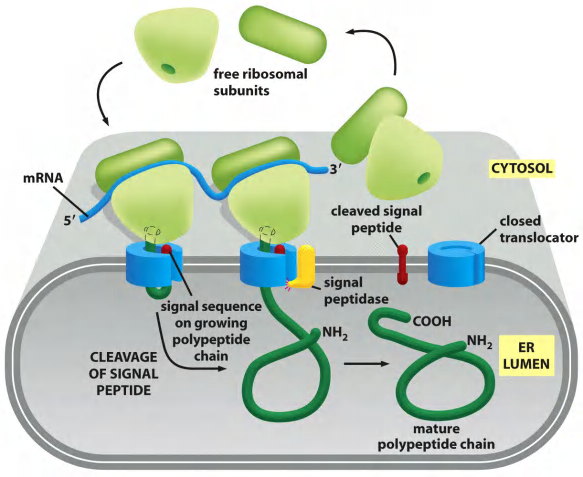
In co-translational transport, the ribosome that is synthesizing the protein is attached directly to the ER membrane, enabling one end of the protein to be translocated into the ER while the rest of the polypeptide chain is being synthesized.
Features
- Membrane-bound ribosomes form the rough ER (free ribosomes are in cytosol).
- If the protein contain SRP (signal-recognition particle), SRP receptor on ER membrane will bind ribosome.
- New protein will move into ER lumen through translocation channel if it has one signal sequence.
The ER Transport Mechanism
Areas of smooth ER from which transport vesicles carrying newly synthesized proteins and lipids bud off for transport to the Golgi apparatus are called transitional ER.
- In certain specialized cells, the smooth ER is abundant and has additional functions
1. The ER import signals
The ER captures selected proteins from the cytosol as they are being synthesized. These proteins are of two types:
- transmembrane proteins, which are only partly translocated across the ER membrane and become embedded in it,
- water-soluble proteins, which are fully translocated across the ER membrane and are released into the ER lumen.
All of these proteins, regardless of their subsequent fate, are directed to the ER membrane by an ER signal sequence, which initiates their translocation by a common mechanism.
According to the signal hypothesis, the size difference reflects the initial presence of a signal sequence that directs the secreted protein to the ER membrane and is then cleaved off by a signal peptidase in the ER membrane before the polypeptide chain has been completed.
ER signal sequences vary greatly in amino acid sequence, but each has eight or more nonpolar amino acids at its center

2. SRP and SRP Receptor Involving Mechanism
Signal peptide is recognized by signal recognition particle (SRP)
The ER signal sequence is guided to the ER membrane by at least two components: a signal-recognition particle (SRP), which cycles between the ER membrane and the cytosol and binds to the signal sequence, and an SRP receptor in the ER membrane
While the SRP and SRP receptor have fewer subunits in bacteria, homologs are present in all cells, indicating that this protein-targeting mechanism arose early in evolution and has been conserved.
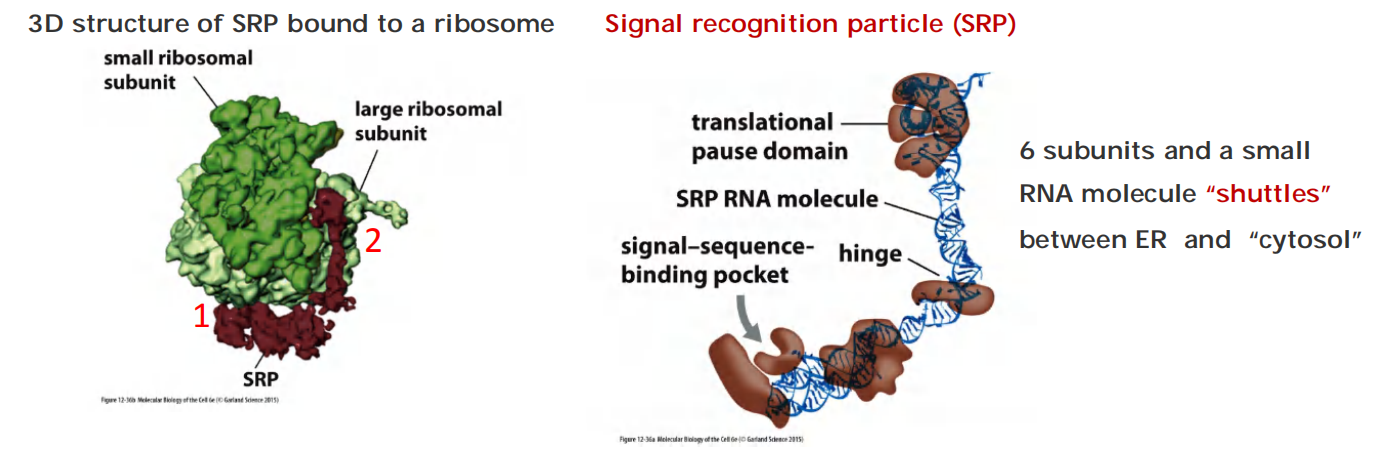
The signal-sequence-binding site is a large hydrophobic pocket lined by methionines. Because methionines have unbranched, flexible side chains, the pocket is sufficiently plastic to accommodate hydrophobic signal sequences of different sequences, sizes, and shapes.
The SRP is a rod-like structure, which wraps around the large ribosomal subunit, with one end binding to the ER signal sequence as it emerges from the ribosome as part of the newly made polypeptide chain; the other end blocks the elongation
Structure evidence:
- SRP binds to the large ribosomal subunit so that its signal-sequence-binding pocket is positioned near the growing polypeptide chain exit site.
- Its translational pause domain is positioned at the interface between the ribosomal subunits, where it interferes with elongation factor binding.
Route
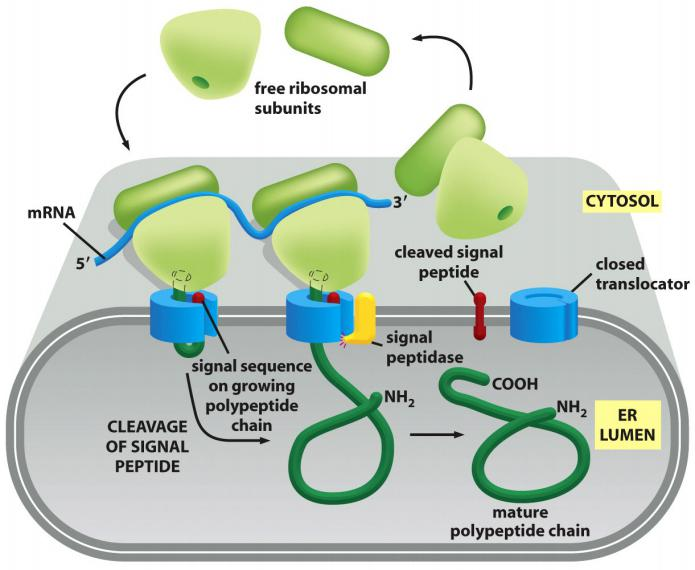
The ribosome binds to mRNA:
Scenario
- Protein translation starts on the free ribosomes in the cytosol.
- Once the N-terminal ER signal peptide emerges, a signal recognition particle (SRP) stops the translation and guides the complex to the translocation pore on the ER, then, translation continues
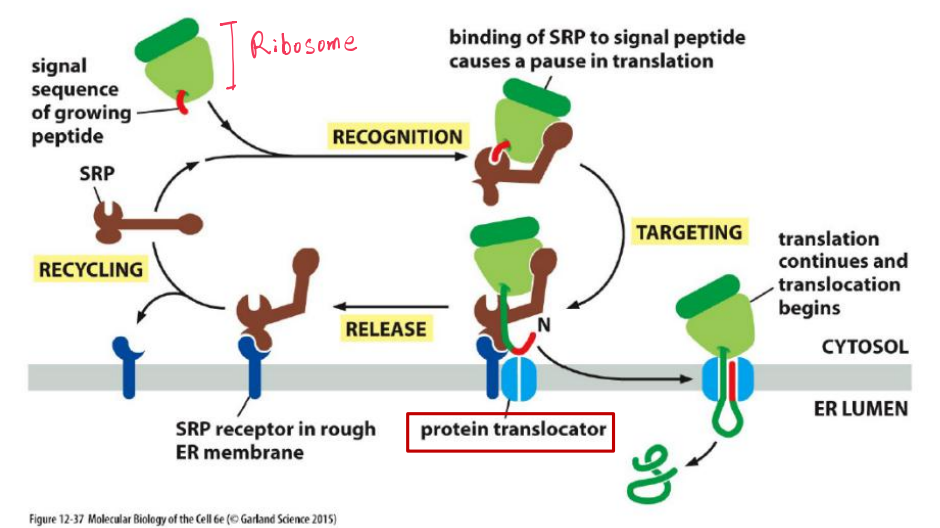
Polypeptide chains passes through an aqueous channel (translocator) in the membrane
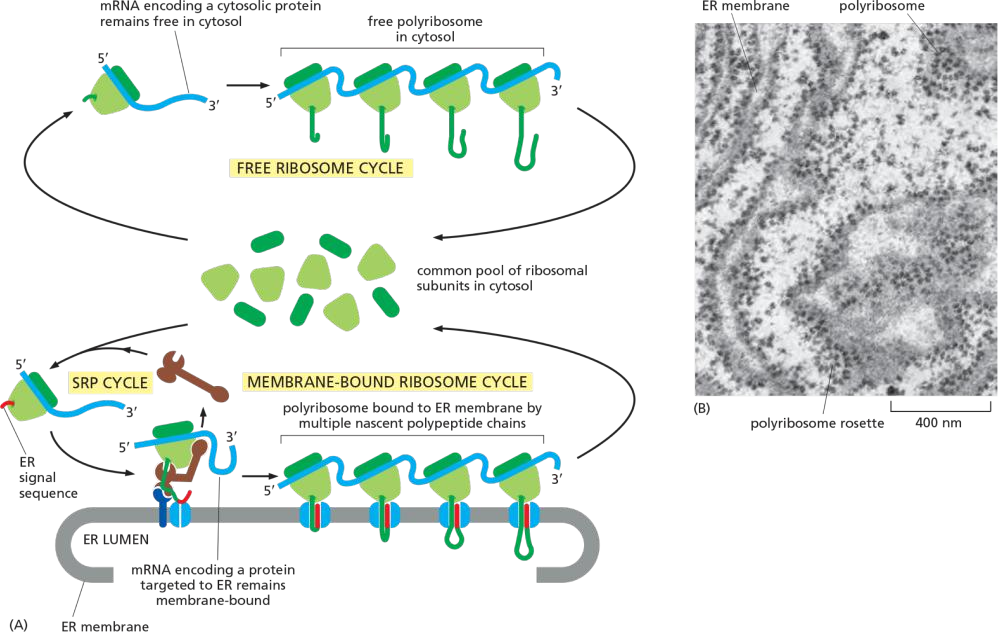
Most proteins imported co-translationally(同时翻译和转运) into the ER through the translocator.
This co-translational transfer process creates two spatially separate populations of ribosomes in the cytosol.
Membrane-bound ribosomes, attached to the cytosolic side of the ER membrane, are engaged in the synthesis of proteins that are being concurrently translocated into the ER
Free ribosomes, unattached to any membrane, synthesize all other proteins encoded by the nuclear genome. Membrane-bound and free ribosomes are structurally and functionally identical.
They differ only in the proteins they are making at any given time.
(1) Polyribosome
Since many ribosomes can bind to a single mRNA molecule, a polyribosome is usually formed. If the mRNA encodes a protein with an ER signal sequence, the polyribosome becomes attached to the ER membrane, directed there by the signal sequences on multiple growing polypeptide chains.
The individual ribosomes associated with such an mRNA molecule can return to the cytosol when they finish translation and intermix with the pool of free ribosomes.
The mRNA itself, however, remains attached to the ER membrane by a changing population of ribosomes, each transiently held at the membrane by the translocator
Protein translocator: Sec61
The Polypeptide Chain Passes Through an Aqueous Channel in the Translocator
The identification of the translocator, which was shown to form a water-filled channel in the membrane through which the polypeptide chain passes. The core of the translocator, called the Sec61 complex, is built from three subunits that are highly conserved from bacteria to eukaryotic cells.
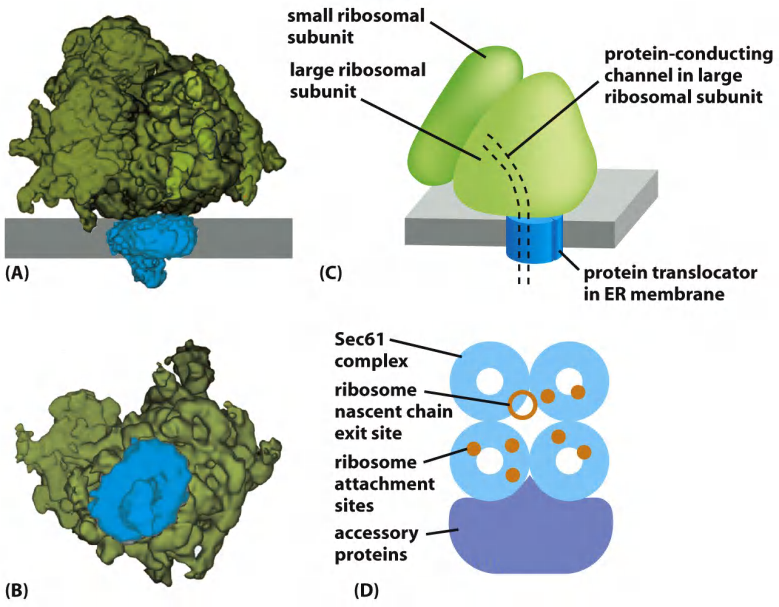
Ribosomes bind to the translocation pore
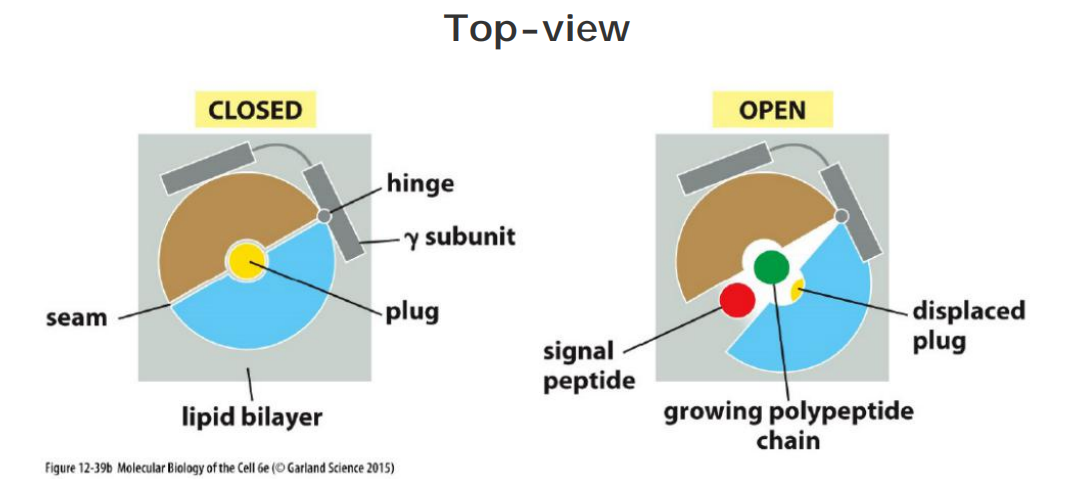
(1) Structure and Gated Channel
The Sec61α subunit has an inverted repeat structure
The structure of the Sec61 complex suggests that α helices contributed by the largest subunit surround a central channel through which the polypeptide chain traverses the membrane
Features
“Closed conformation”: the yellow short helix is thought to form a plug(塞子) that seals the pore when the translocator is closed. (Showed in the figure below.)
To open, the complex rearranges itself thereby moving the plug helix out of the way (red arrow).
Prevent leaking: a ring of hydrophobic amino acid side chains is thought to form a tight-fitting diaphragm around the translocating polypeptide chain to prevent leaks of other molecules across the membrane.
The pore of the Sec61 complex can also open sideways at a lateral seam.
Model of the translocator function
The short helix which showed in yellow color below is thought to form a plug that seals the pore when the translocator is closed
The channel is gated by a short α helix that is thought to keep the translocator closed when it is idle and to move aside when it is engaged in passing a polypeptide chain. According to this view, the pore is a dynamic gated channel that opens only transiently when a polypeptide chain traverses the membrane
The structure of the Sec61 complex suggests that the pore can also open along a seam on its side
- This opening allows a translocating peptide chain lateral access into the hydrophobic core of the membrane, a process that is important both for the release of a cleaved signal peptide into the membrane
In eukaryotic cells, four Sec61 complexes form a large translocator assembly that can be visualized on ER-bound ribosomes after detergent solubilization of the ER membrane
- It is likely that this assembly includes other membrane complexes that associate with the translocator, such as enzymes that modify the growing polypeptide chain, including oligosaccharide transferase and the signal peptidase. The assembly of a translocator with these accessory components is called the translocon.
Structurally similar translocators
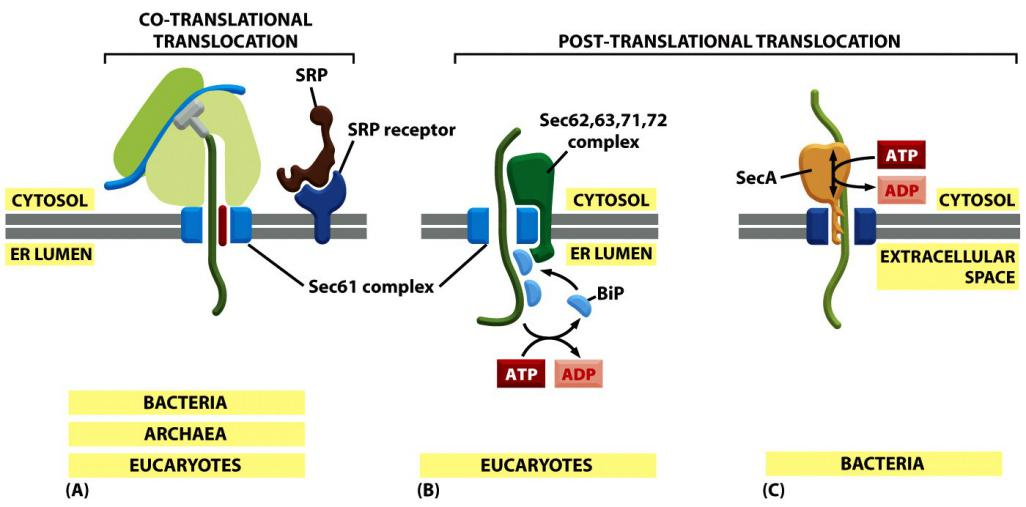
Translocation does not always require ongoing protein elongation.
In bacteria, a translocation motor protein, the SecA ATPase, attaches to the cytosolic side of the translocator, where it undergoes cyclic conformational changes driven by ATP hydrolysis
- Each time an ATP is hydrolyzed, a portion of the SecA protein inserts into the pore of the translocator, pushing a short segment of the passenger protein with it
- As a result of this ratchet((防倒转的)棘齿) mechanism, the SecA ATPase progressively pushes the polypeptide chain of the transported protein across the membrane.
Eukaryotic cells use a different set of accessory proteins that associate with the Sec61 complex.
- These proteins span the ER membrane and use a small domain on the lumenal side of the ER membrane to deposit an hsp70-like chaperone protein (called BiP, for binding protein) onto the polypeptide chain as it emerges from the pore into the ER lumen
- ATP-dependent cycles of BiP binding and release drive unidirectional translocation, as described earlier for the mitochondrial hsp70 proteins that pull proteins across mitochondrial membranes.
Proteins that are transported into the ER by a post-translational mechanism are first released into the cytosol, where they bind to chaperone proteins to prevent folding, as discussed earlier for proteins destined for mitochondria and chloroplasts
Transport into the ER: co- and post—translational translocation
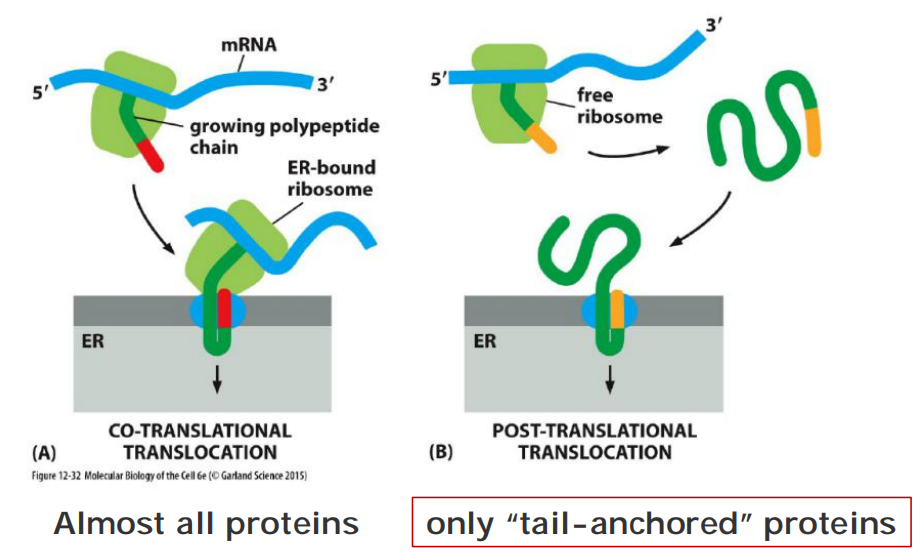
Some completely synthesized proteins, however, are imported into the ER, demonstrating that translocation does not always require ongoing translation
In contrast to the post-translational transport into the nucleus, mitochondria, chloroplasts and peroxisomes.
Co-translational translocation of almost all ER proteins (not all, tail-anchored proteins use the post-translational mechanism)
Double Check for The Protein import into ER
An ER signal sequence is therefore recognized twice:
- first by an SRP in the cytosol and then by a binding site in the pore of the protein translocator, where it serves as a start-transfer signal (or start-transfer peptide) that opens the pore (for example, see Figure 12–35 for how this works for a soluble protein)
Dual recognition may help ensure that only appropriate proteins enter the lumen of the ER.
When the nascent(初期的;开始存在的) polypeptide chain grew long enough, the ER signal peptidase cleaved off the signal sequence and released it from the pore into the membrane, where it was rapidly degraded to amino acids by other proteases in the ER membrane
Double-check for the proteins to be transported by signal sequence
- binding to SRP
- binding to translocator
3. Different Kinds of Proteins import into ER
Translocation of proteins into ER do differ, which is dependent on their topology
Proteins differ:
- Water-soluble proteins :
- ER lumen resident protein
- proteins to be transported Golgi, endosome, and lysosome lumen
- proteins to be secreted
The way how a protein is translocated into the endoplasmic reticulum (ER) depends on the topology of the respective proteins
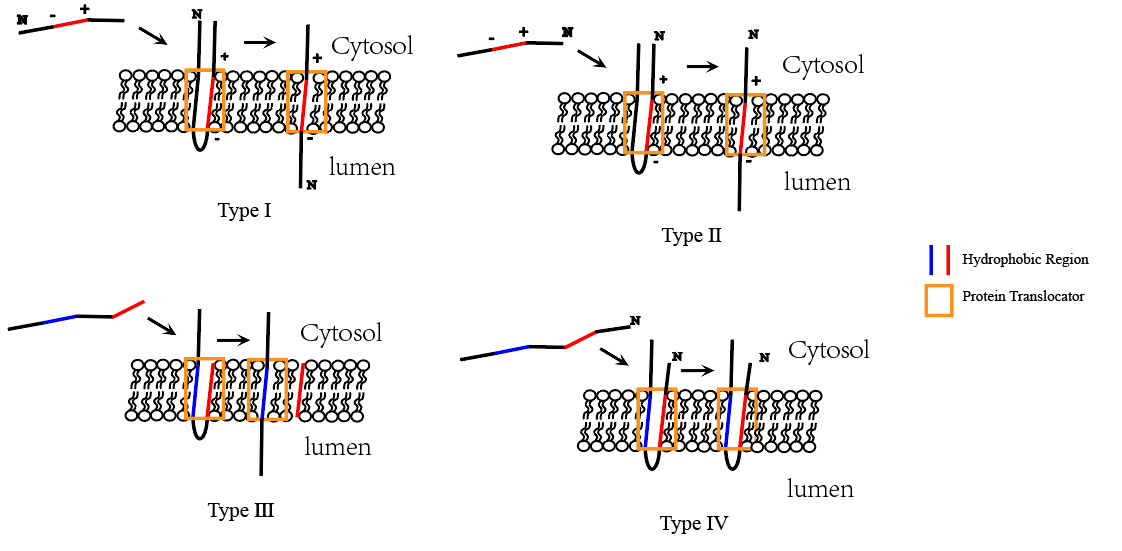
Single-pass transmembrane protein types
- Type I transmembrane proteins: N-terminus in the ER lumen, C-terminus in the cytosol.
- Type II transmembrane proteins: C-terminus in the ER lumen, N-terminus in the cytosol.
(1) Translocation of water-soluble proteins to the ER lumen
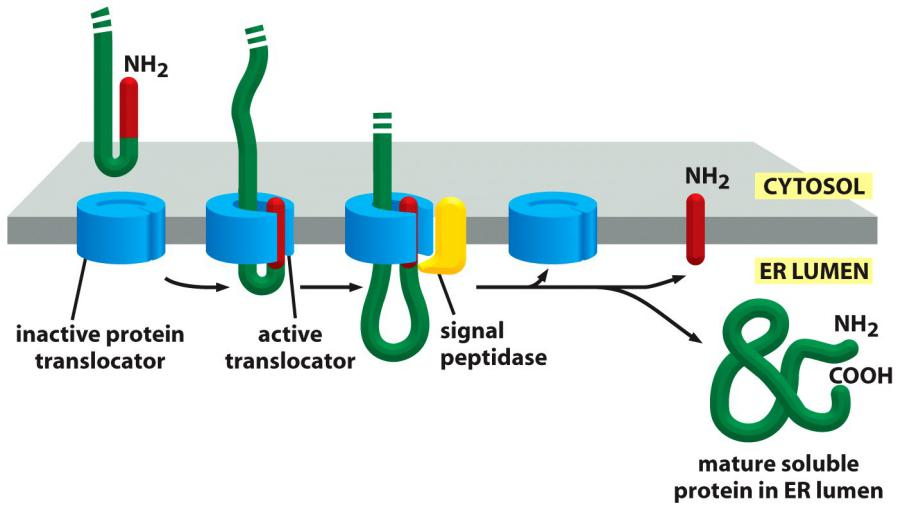
The N-terminal signal sequence (signal peptide) acts as start-transfer sequence, it is “opening the pore”
This signal peptide left in the ER membrane cleaved off by the enzyme signal peptidase immediately after translocation
(2) Single-pass integral membrane proteins contain stop transfer sequence
In Single-Pass Transmembrane Proteins, a Single Internal ER Signal Sequence Remains in the Lipid Bilayer as a Membrane-spanning α Helix
The ER signal sequence in the growing polypeptide chain is thought to trigger the opening of the pore in the Sec61 protein translocator: after the signal sequence is released from the SRP and the growing chain has reached a sufficient length, the signal sequence binds to a specific site inside the pore itself, thereby opening the pore
Insertion via N-terminal start-transfer signal and a stop-transfer signal
In the simplest case, an N-terminal signal sequence initiates translocation, just as for a soluble protein, but an additional hydrophobic segment in the polypeptide chain stops the transfer process before the entire polypeptide chain is translocated. This stop-transfer signal anchors the protein in the membrane after the ER signal sequence (the start-transfer signal) has been cleaved off and released from the translocator
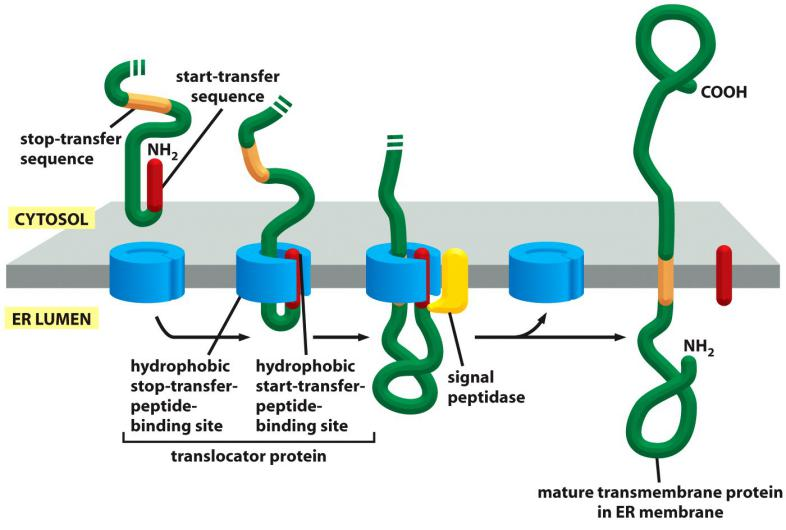
Prerequisites for the insertion:
- Recognition of a N-terminal signal peptide for the ER by the SRP: start-transfer signal
- Recognition of the Internal hydrophobic peptide the translocator: stop-transfer signal
- The start-transfer signal is cleaved off and the hydrophobic “stop-transfer” signal anchors the protein.
Single-pass integral membrane proteins have different topology
In the other two cases, the signal sequence is internal, rather than at the N-terminal end of the protein. As for an N-terminal ER signal sequence, the SRP binds to an internal signal sequence by recognizing its hydrophobic α-helical features.
After release from the translocator, the internal start-transfer sequence remains in the lipid bilayer as a single membrane-spanning α helix
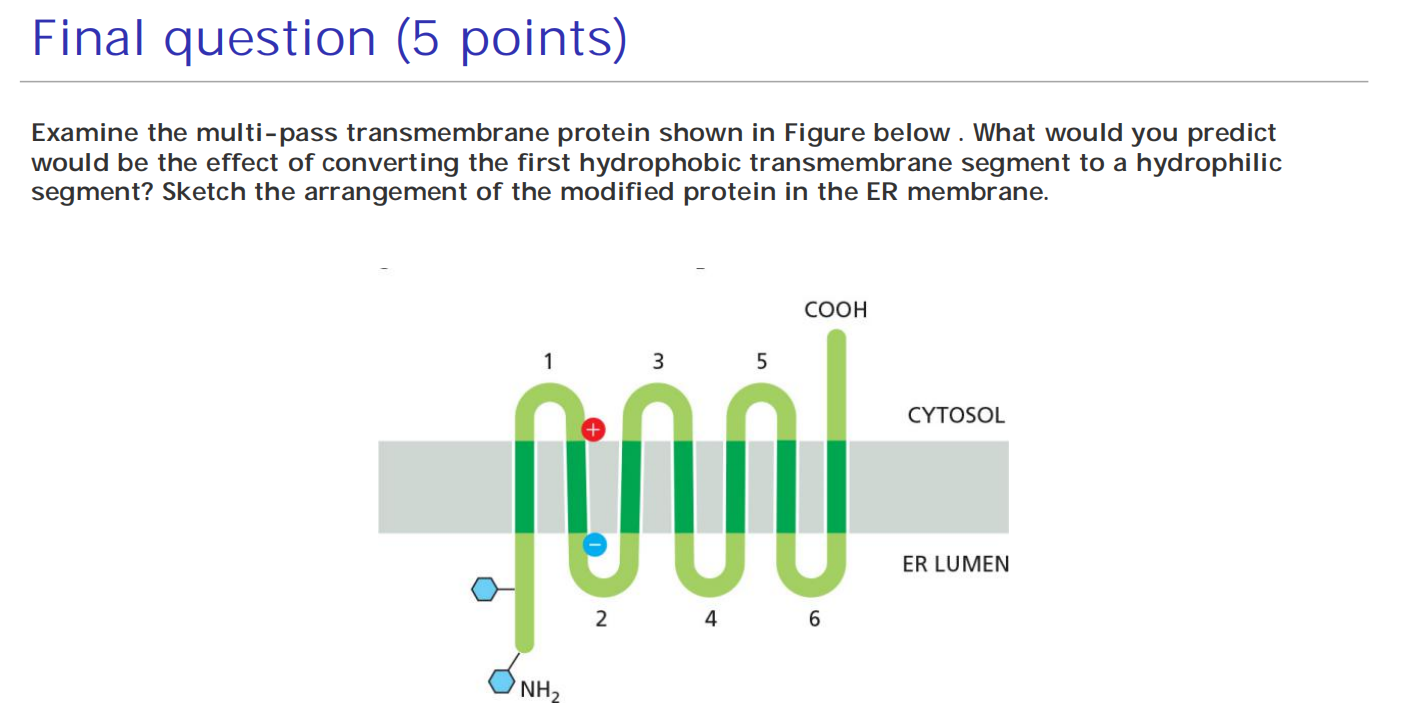
The orientation of the start-transfer sequence depends on the distribution of nearby charged amino acids,
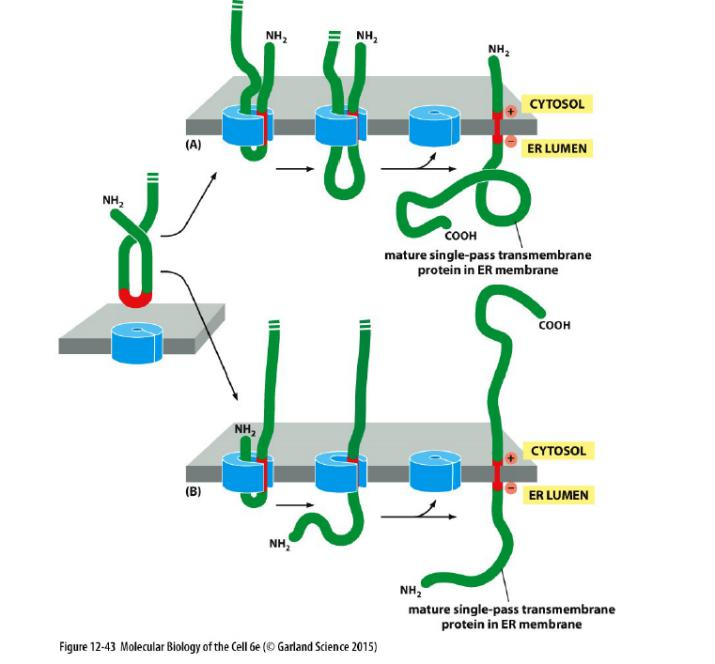
Charge of amino acids that flanks the internal start-transfer sequence can change the topology
- Type II configuration (C-terminus in the ER lumen, N-terminus in the cytosol) (consider the topology)
- Positively charged amino acid before the internal start-transfer sequence!
Configuration depends on the charge of flanking amino acids.
- Type I configuration (N-terminus in the ER lumen, C-terminus in the cytosol) (consider the topology)
- Positively charged amino acids after the internal start-transfer sequence!
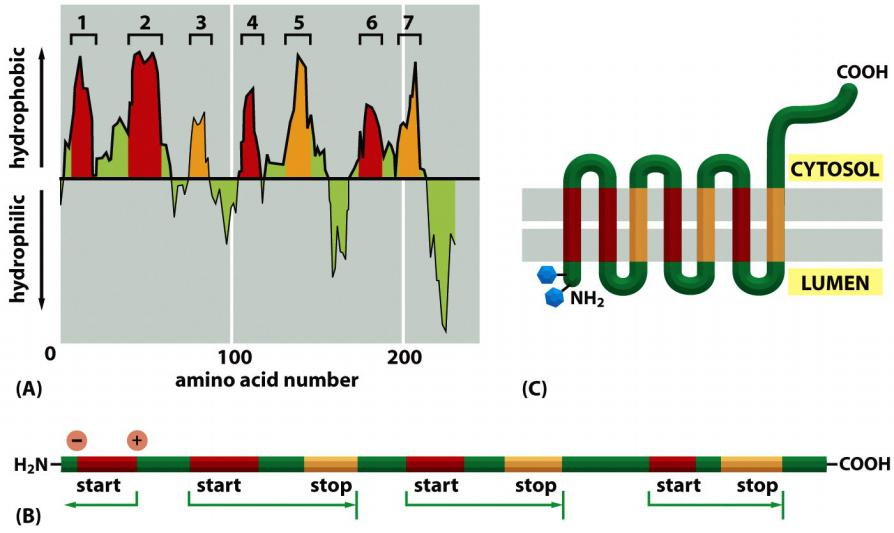
Multi-pass transmembrane proteins
Combinations of Start-Transfer and Stop-Transfer Signals Determine the Topology of Multipass Transmembrane Proteins
In multipass transmembrane proteins, the polypeptide chain passes back and forth repeatedly across the lipid bilayer as hydrophobic α helices
- It is thought that an internal signal sequence serves as a start-transfer signal in these proteins to initiate translocation, which continues until the translocator encounters a stop-transfer sequence
(1) Insertion of a double-pass transmembrane protein into the ER
Two signals: an internal start-transfer signal and a stop-transfer signal
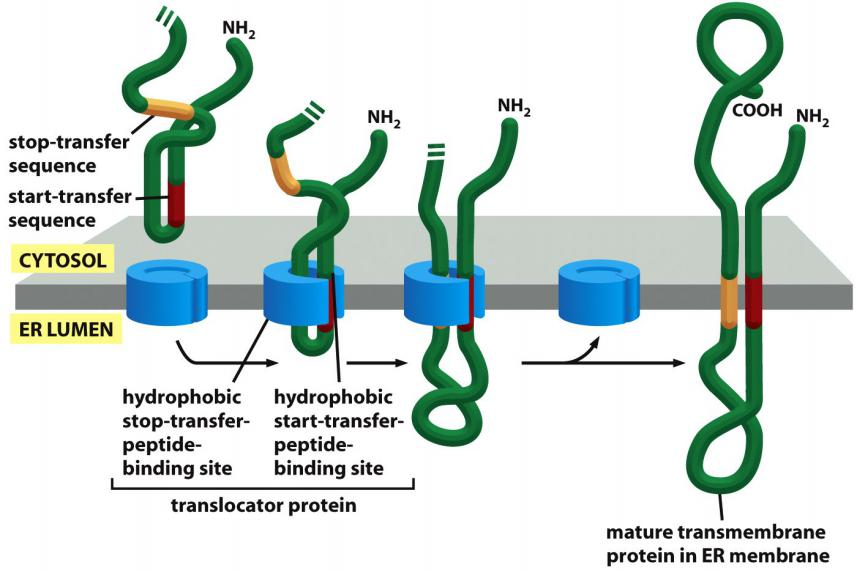
(2) Multiple pairs of “start-transfer” and “stop-transfer” signals
In more complex multipass proteins, in which many hydrophobic α helices span the bilayer, a second start-transfer sequence reinitiates translocation further down the polypeptide chain until the next stop-transfer sequence causes polypeptide release, and so on for subsequent start-transfer and stop-transfer sequences
Whether a given hydrophobic signal sequence functions as a start-transfer or stop-transfer sequence must depend on its location in a polypeptide chain,
- Thus, the distinction between start-transfer and stop-transfer sequences results mostly from their relative order in the growing polypeptide chain
- It seems that the SRP begins scanning an unfolded polypeptide chain for hydrophobic segments at its N-terminus and proceeds toward the C-terminus, in the direction that the protein is synthesized.
- A similar scanning process continues until all of the hydrophobic regions in the protein have been inserted into the membrane as transmembrane α helices
The way in which a newly synthesized protein is inserted into the ER membrane determines the orientation of the protein in all of the other membranes as well.
Tail-anchored proteins
ER Tail-anchored Proteins Are Integrated into the ER Membrane by a Special Mechanism
- Many important membrane proteins are anchored in the membrane by a C-terminal transmembrane, hydrophobic α helix
These ER tail-anchored proteins include a large number of SNARE protein subunits that guide vesicular traffic
ER tail-anchored protein is integrated by a special mechanism
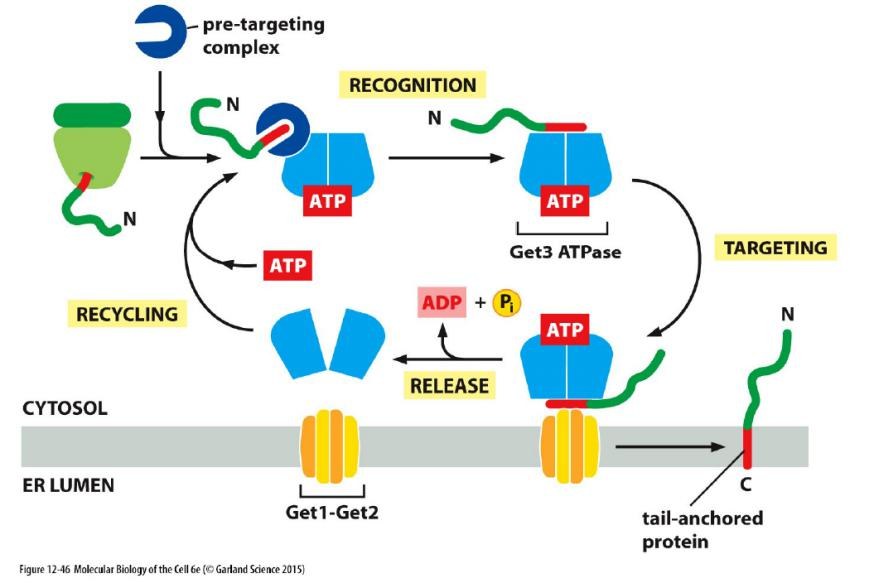
When such a tail-anchored protein inserts into the ER membrane from the cytosol, only a few amino acids that follow the transmembrane α helix on its C-terminal side are translocated into the ER lumen, while most of the protein remains in the cytosol
Because of the unique position of the transmembrane α helix in the protein sequence, translation terminates while the C-terminal amino acids that will form the transmembrane α helix have not yet emerged from the ribosome exit tunnel
Recognition by SRP is therefore not possible. It was long thought that these proteins are released from the ribosome and the hydrophobic C-terminal tail spontaneously partitions into the ER membrane.
It is now clear that a specialized targeting machinery is involved that is fueled by ATP hydrolysis
- Although the components and details differ, this post-translational targeting mechanism is conceptually similar to SRP-dependent protein targeting
Protein folding and “quality control” in the ER
Features
Translocated polypeptide chains are folded and assembled in the ER lumen.
Most proteins in rough ER are glycosylated via the addition of a common N-linked oligosaccharide. (∼ 50% of the soluble and membrane-bound proteins are glycosylated)
For details, see lecture 12
- Protein folding is influenced by chaperons
- Quality control is associated with Glycosylation
Many of the proteins in the lumen of the ER are in transit, en route (在途中) to other destinations; others, however, normally reside there and are present at high concentrations.
These ER resident proteins contain an ER retention signal of four amino acids at their C-terminus that is responsible for retaining the protein in the ER
- Some of these proteins function as catalysts that help the many proteins that are translocated into the ER lumen to fold and assemble correctly.
One important ER resident protein is protein disulfide isomerase (PDI), which catalyzes the oxidation of free sulfhydryl (SH) groups on cysteines to form disulfide (S–S) bonds
- Almost all cysteines in protein domains exposed to either the extracellular space or the lumen of organelles in the secretory and endocytic pathways are disulfide-bonded
- By contrast, disulfide bonds form only very rarely in domains exposed to the cytosol, because of the reducing environment there
Another ER resident protein is the chaperone protein BiP. We have already discussed how BiP pulls proteins post-translationally into the ER through the Sec61 ER translocator.
- Like other chaperones, BiP recognizes incorrectly folded proteins, as well as protein subunits that have not yet assembled into their final oligomeric complexes
- The bound BiP both prevents the protein from aggregating and helps keep it in the ER (and thus out of the Golgi apparatus and later parts of the secretory pathway).
- Like some other members of the hsp70 family of chaperone proteins, which bind unfolded proteins and facilitate their import into mitochondria and chloroplasts, BiP hydrolyzes ATP to shuttle between high- and low-affinity binding states, which allow it to hold on to and let go of its substrate proteins in a dynamic cycle
二、Glycosylation: sweet life
Glycosylation is add sugar molecule to proteins.
Most Proteins Synthesized in the Rough ER Are Glycosylated by the Addition of a Common N-Linked Oligosaccharide
- The covalent addition of oligosaccharides to proteins is one of the major biosynthetic functions of the ER
Function of glycosylation
- Folding
- Transport (e. g. sorting, M6P-receptor)
- Stability (quality control)
- Recognition (e.g. lectin)
- Regulatory roles (e.g. Notch protein)
Two types of glycosylation
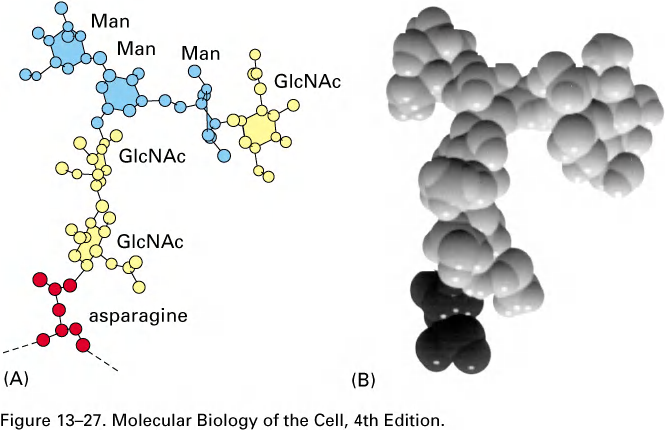
During the most common form of protein glycosylation in the ER, a preformed precursor oligosaccharide (composed of N-acetylglucosamine, mannose, and glucose, and containing a total of 14 sugars) is transferred en bloc (整块) to proteins.
- Because this oligosaccharide is transferred to the side-chain hemiacetal group (NH2) of an asparagine in the protein, it is said to be N-linked or asparagine-linked
- The N-linked oligosaccharides are by far the most common oligosaccharides, being found on 90% of all glycoproteins
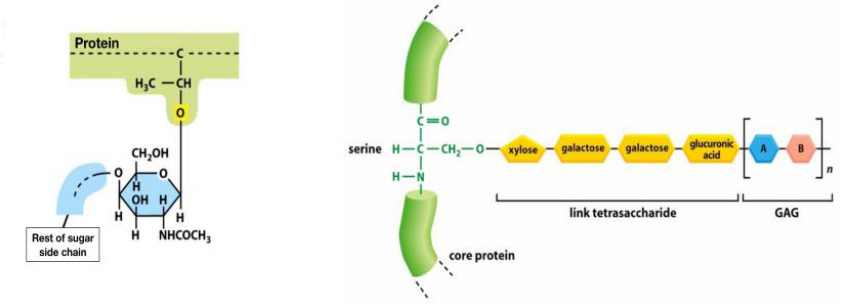
Less frequently, oligosaccharides are linked to the hydroxyl group(羟基,-OH) on the side chain of a serine (S), threonine (T), or hydroxylysine(羟(基)赖氨酸) amino acid. They are called O-linked oligosaccharides.
- A first sugar of these O-linked oligosaccharides is added in the ER and the oligosaccharide is then further extended in the Golgi apparatus.(在高尔基体中进一步得到扩展)
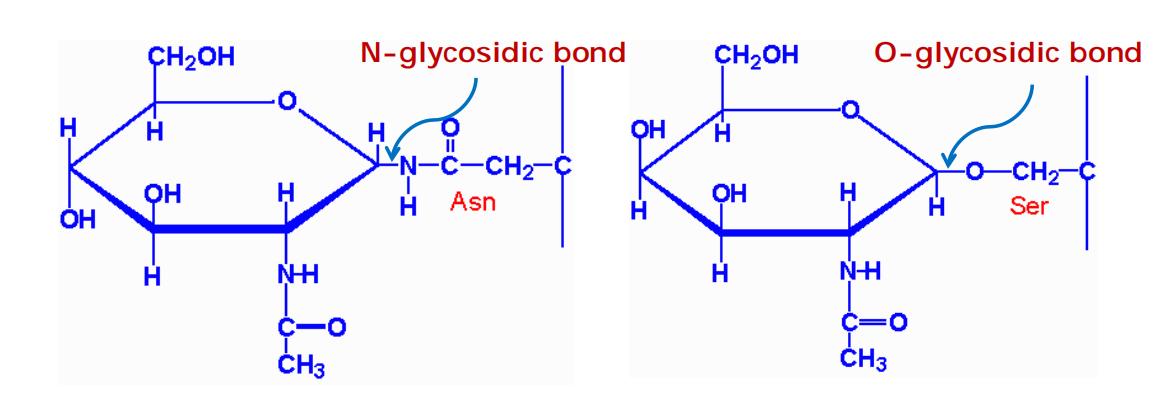
N-glycosidic bonds form via an N-glycosidic linkage is through the amide group of asparagine and the hemiacetal group (NH
2) of N-acetylglucosamineO-glycosidic bonds form via an O-glycosidic linkage is through the hydroxyl group (OH) of serine and the hemiacetal group of N-acetylglucosamine
1. Two main classes of N-linked glycosylation
- In the ER
- In the Golgi
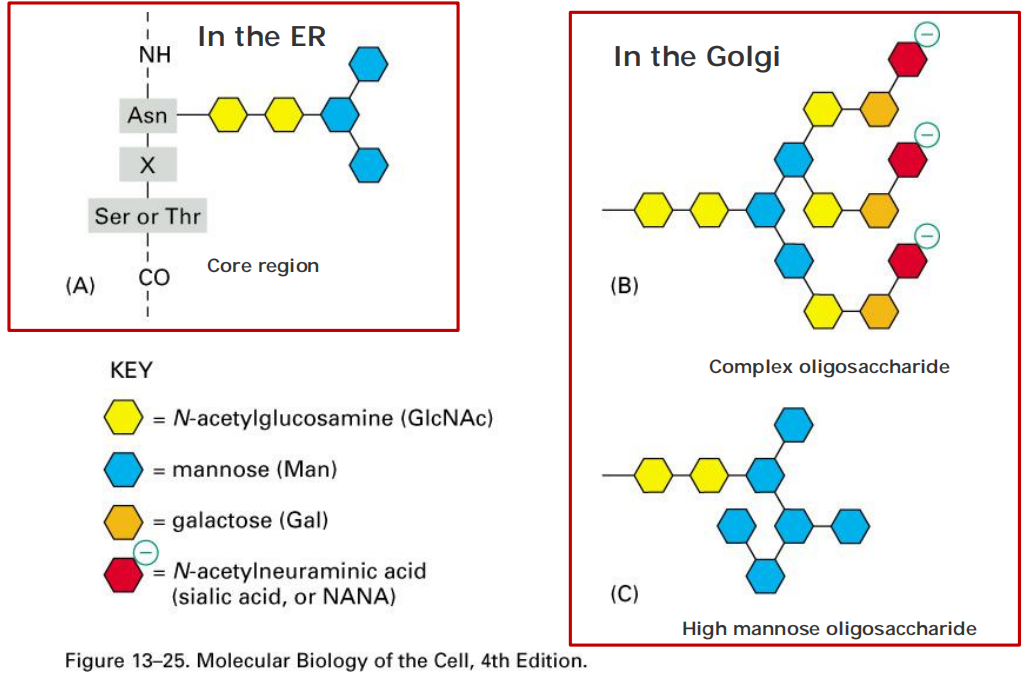
Glycosylation occurs in the ER and Golgi
1. Protein glycosylation in the ER
The precursor oligosaccharide is built up sugar by sugar on the membrane-bound dolichol lipid(多元醇脂质) and is then transferred to a protein.
The sugars are first activated in the cytosol by the formation of nucleotide (UDP or GDP)-sugar intermediates, which then donate their sugar (directly or indirectly) to the lipid in an orderly sequence
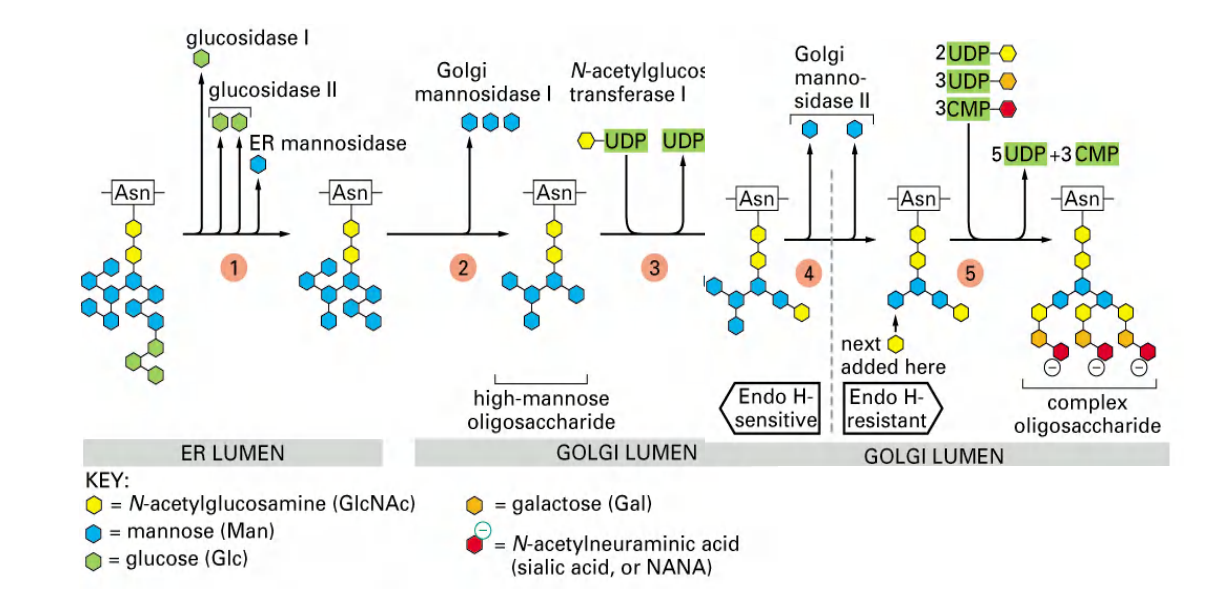
All of the diversity of the N-linked oligosaccharide structures on mature glycoproteins results from the later modification of the original precursor oligosaccharide.
- While still in the ER, three glucoses (see Figure 12–47) and one mannose are quickly removed from the oligosaccharides of most glycoproteins. We shall return to the importance of glucose trimming shortly
- This oligosaccharide“trimming,” or“processing,”(修饰)continues in the Golgi apparatus
Protein glycosylation in the ER: N (asparagine)-linked glycosylation
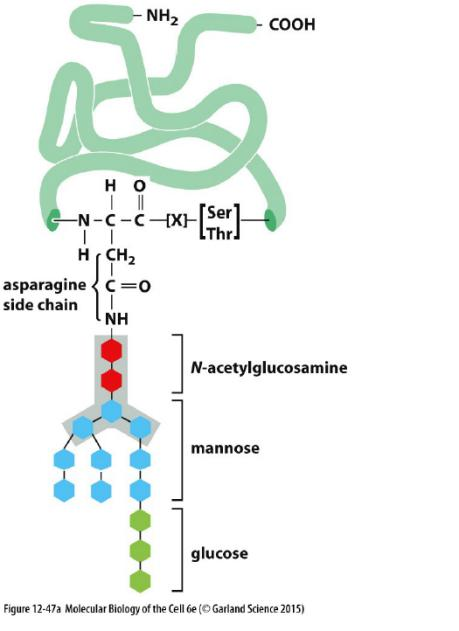
- N-linked oligosaccharides takes up 90% of all glyco-proteins.
The precursor oligosaccharide is attached only to asparagine in the sequence Asn-X-Ser and Asn-X-Thr (where X is any amino acid except proline (For proline has bulk side chain)).
The other from O-linked in Golgi apparatus (Ser, Thr, hydroxylyserine).
Oligosaccharides are used as tags to mark the state of protein folding in the ER
A clue to the role of glycosylation in protein folding came from studies of two ER chaperone proteins, which are called calnexin(钙结合蛋白) and calreticulin(钙网蛋白) because they require Ca2+ for their activities
- These chaperones are carbohydrate-binding proteins, or lectins(凝集素), which bind to oligosaccharides on incompletely folded proteins and retain them in the ER
- Like other chaperones, they prevent incompletely folded proteins from irreversibly aggregating
Quality control of protein folding: calnexin (钙黏连蛋白) in and glucosyl transferase(葡萄糖基转移酶) prevent unfolded proteins from leaving the ER
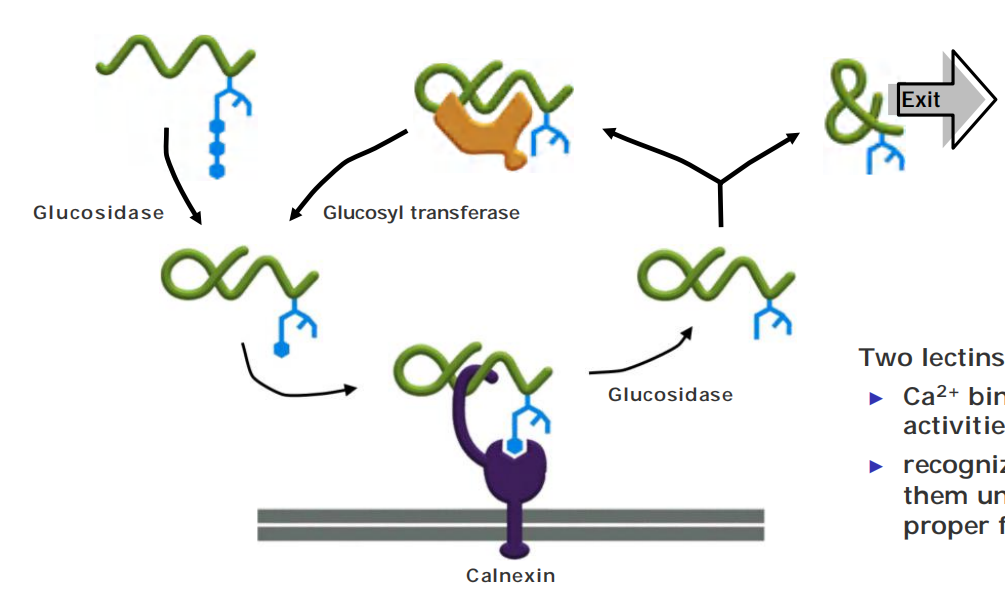
Two lectins: calnexin (钙联结蛋白) & calreticulin (钙网蛋白)
- Ca2+ binding essential for their activities
- recognize sugar group and bind them until unfolded protein is proper folded and exits ER
When the third glucose has been removed, the glycoprotein dissociates from its chaperone and can leave the ER.
How, then, do calnexin and calreticulin distinguish properly folded from incompletely folded proteins? The answer lies in yet another ER enzyme, a glucosyl transferase that keeps adding a glucose to those oligosaccharides that have lost their last glucose
Mannosidase (甘露糖苷酶) triggers degradation pathway for ER proteins
在一定时间内,不断进行循环,如果蛋白proper folded, 则转运出ER
若始终未能proper folded,则被mannosidase降解。
improper folded protein存在的时间越长,与mannosidase相遇的机会越大。
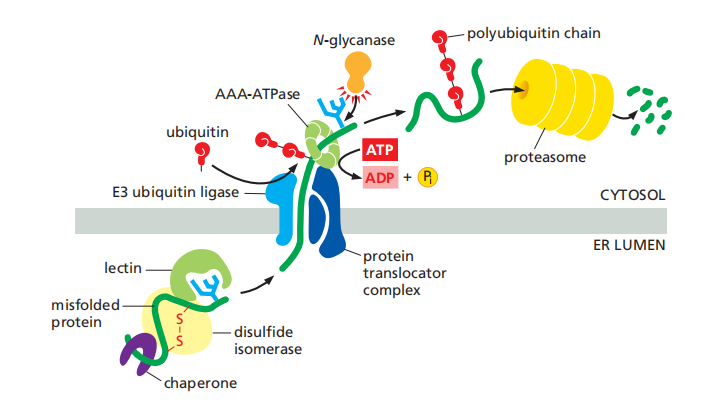
Improperly Folded Proteins Are Exported from the ER and Degraded in the Cytosol
- Such proteins are exported from the ER back into the cytosol, where they are degraded in proteasomes
- The N-linked oligosaccharides serve as timers that measure how long a protein has spent in the ER.
The slow trimming of a particular mannose on the core oligosaccharide tree by an enzyme (a mannosidase) in the ER creates a new oligosaccharide structure that ER-lumenal lectins of the retrotranslocation apparatus recognize.
Multiple translocator complexes move different proteins from the ER membrane or lumen into the cytosol.
A common feature is that they each contain an E3 ubiquitin ligase enzyme, which attaches polyubiquitin tags to the unfolded proteins as they emerge into the cytosol, marking them for destruction.
Fueled by the energy derived from ATP hydrolysis, a hexomeric ATPase of the family of AAA-ATPases
- An N-glycanase removes its oligosaccharide chains en bloc(整块). Guided by its ubiquitin tag, the deglycosylated polypeptide is rapidly fed into proteasomes, where it is degraded
Misfolded Proteins in the ER Activate an Unfolded Protein Response
- An accumulation of misfolded proteins in the cytosol, for example, triggers a heat-shock response, which stimulates the transcription of genes encoding cytosolic chaperones that help to refold the proteins
- Similarly, an accumulation of misfolded proteins in the ER triggers an unfolded protein response, which includes an increased transcription of genes encoding proteins involved in retrotranslocation and protein degradation in the cytosol, ER chaperones, and many other proteins that help to increase the protein-folding capacity of the ER.
Pathways that execute the unfolded protein response
There are three parallel pathways that execute the unfolded protein response
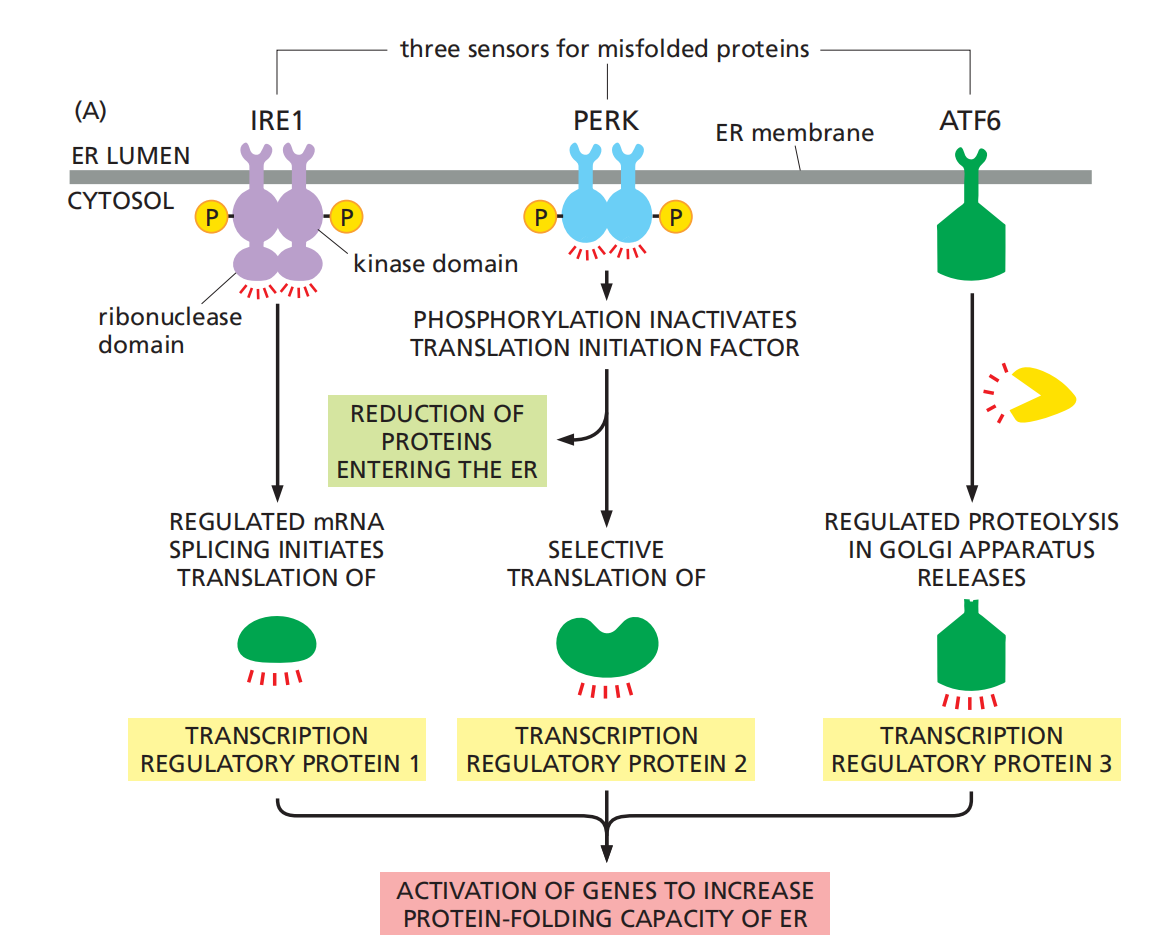
IRE1: The first pathway, which was initially discovered in yeast cells, is particularly remarkable. Misfolded proteins in the ER activate a transmembrane protein kinase in the ER, called IRE1, which causes the kinase to oligomerize and phosphorylate itself.
The oligomerization and autophosphorylation of IRE1 activates an endoribonuclease(核糖核酸内切酶) domain in the cytosolic portion of the same molecule, which cleaves a specific cytosolic mRNA molecule at two positions, excising an intron. This regulates mRNA splicing and finally causes an increase expression of protein chaperon.
PERK: Misfolded proteins also activate a second transmembrane kinase in the ER, PERK, which inhibits a translation initiation factor by phosphorylating it, thereby reducing the production of new proteins throughout the cell.
One consequence of the reduction in protein synthesis is to reduce the flux of proteins into the ER, thereby reducing the load of proteins that need to be folded there.
ATF6: Finally, a third transcription regulator, ATF6, is initially synthesized as a transmembrane ER protein. Because it is embedded in the ER membrane, it cannot activate the transcription of genes in the nucleus.
When misfolded proteins accumulate in the ER, however, the ATF6 protein is transported to the Golgi apparatus, where it encounters proteases that cleave off its cytosolic domain, which can now migrate to the nucleus and help activate the transcription of genes encoding proteins involved in the unfolded protein response.
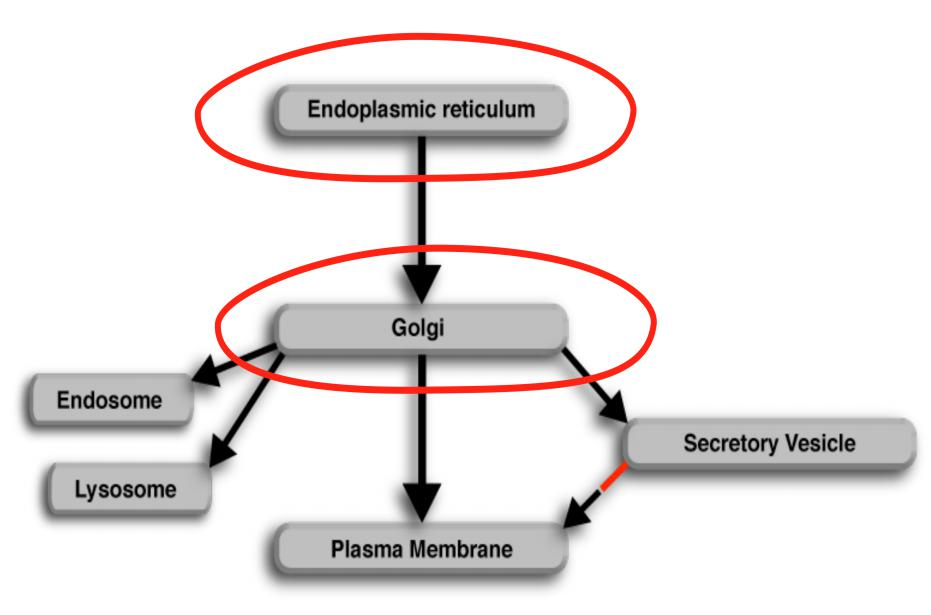
The Golgi apparatus
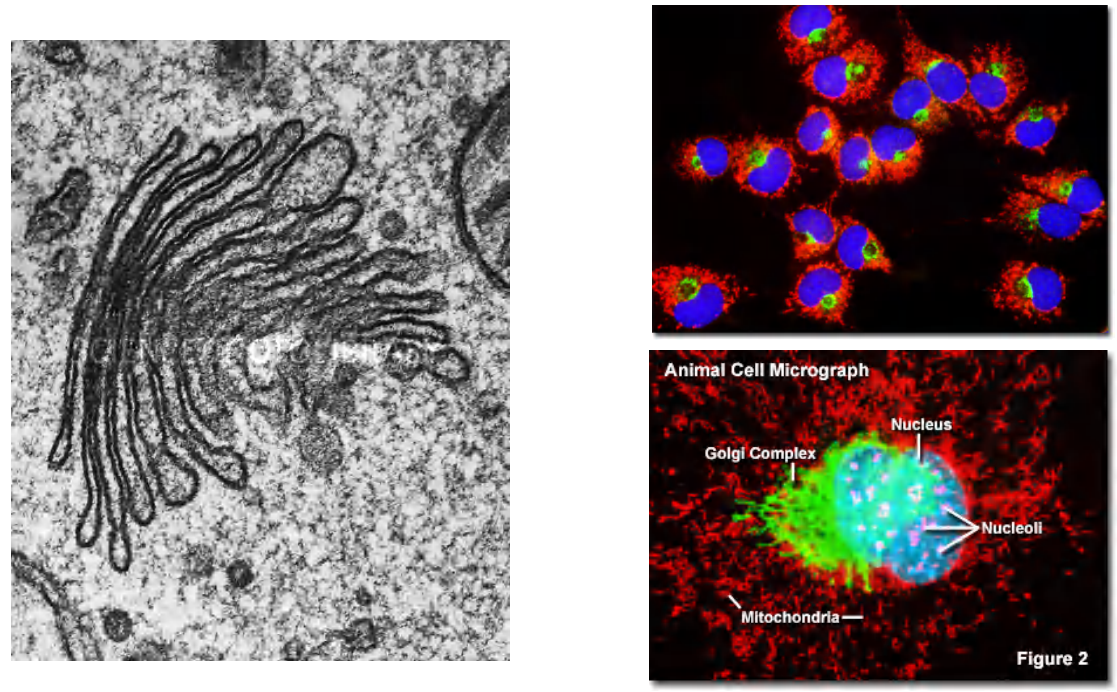
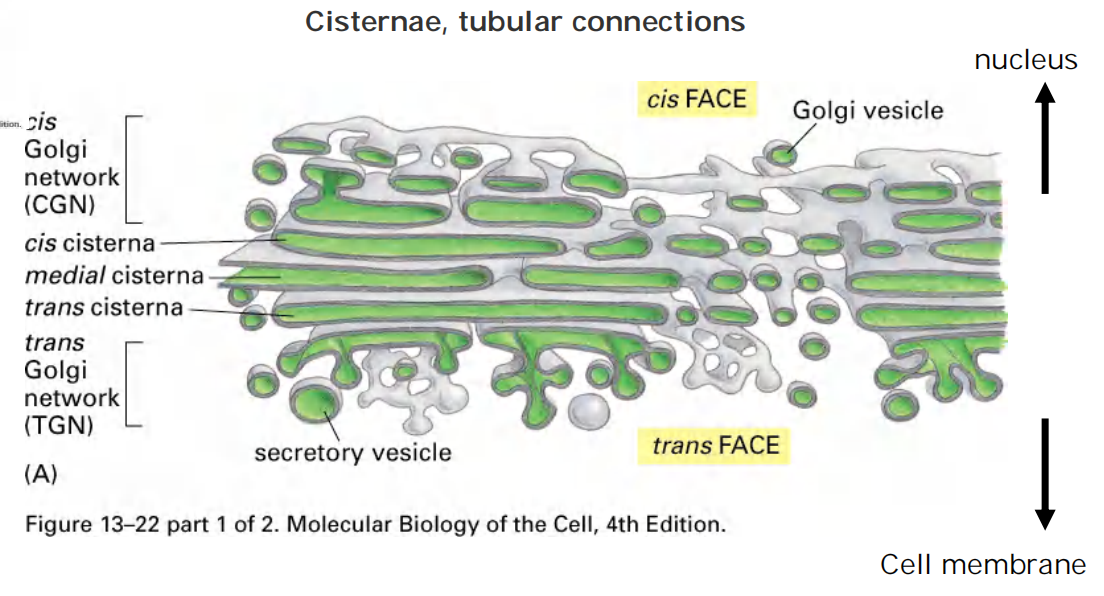
1. Ordered series of Golgi apparatus
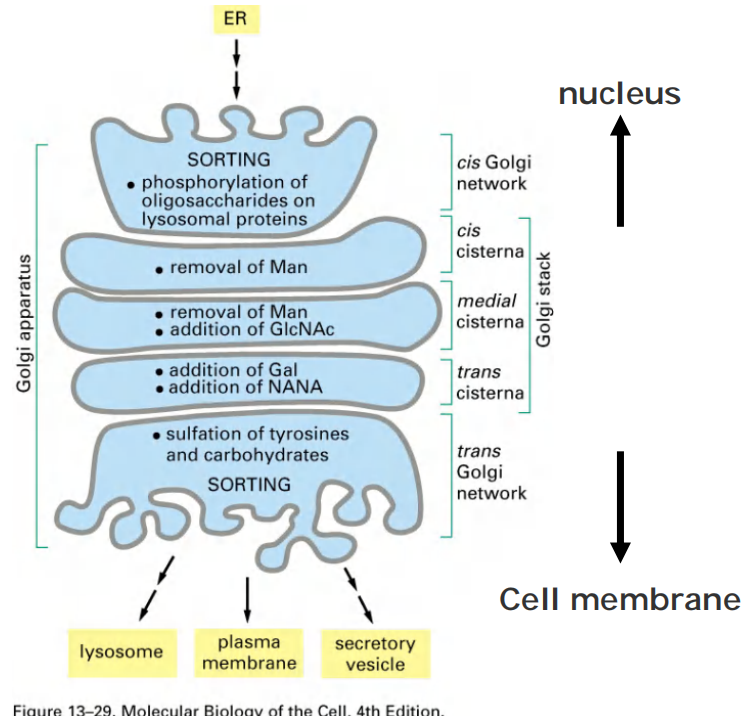
2. Functional compartmentation of Golgi apparatus
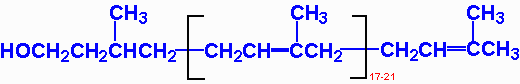
3. Dolichol 长醇
Dolichol: lipid molecules anchor sugar molecules to the phospholipid bilayer.
Protein glycosylation
1. Assembly of the precursor oligosaccharide
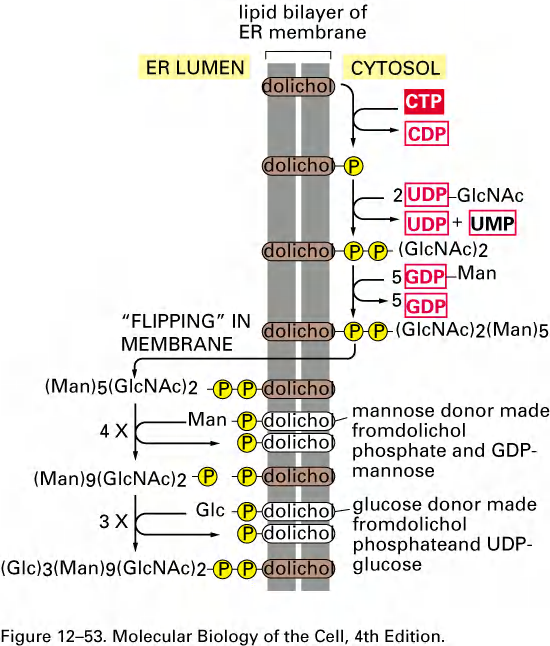
The first step: Synthesis of oligosaccharide
- Assembly takes place on the carrier lipid dolichol, anchored in the ER membrane.
- A pyrophosphate bridge (焦磷酸盐桥) joins the 1st sugar to the dolichol-GDP.
- Sugars are added singly (逐一地) and sequentially.
- After the two N-acetylglucos-amines are added, the assembly flips from the cytosolic side to the ER lumen.
- Nine mannose and three glucose molecules are added, totaling 14 sugars.
- The assembly of sugar molecules starts in the cytosol and is finalized in the ER!
Second step, attachment to protein:
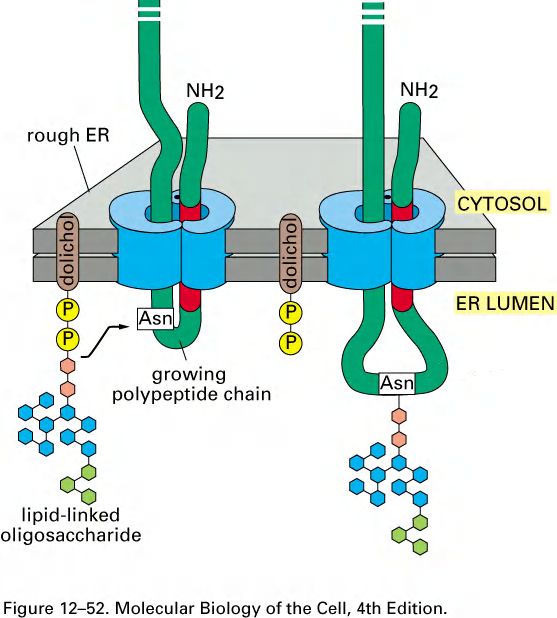
Now in the second step, attachment to protein:
- One-step transfer, catalyzed by oligosaccharyl transferase (寡糖基转移酶), which is bound to the ER membrane next to translocator.
- Covalently attached to certain Asn (Asparagine) in the polypeptide chain (said to be “N-linked” glycosylation).
- Attaches to NH2 side chain of Asn but only in the context:
- Asn-x-Ser or Asn-x-Thr (Threonine)
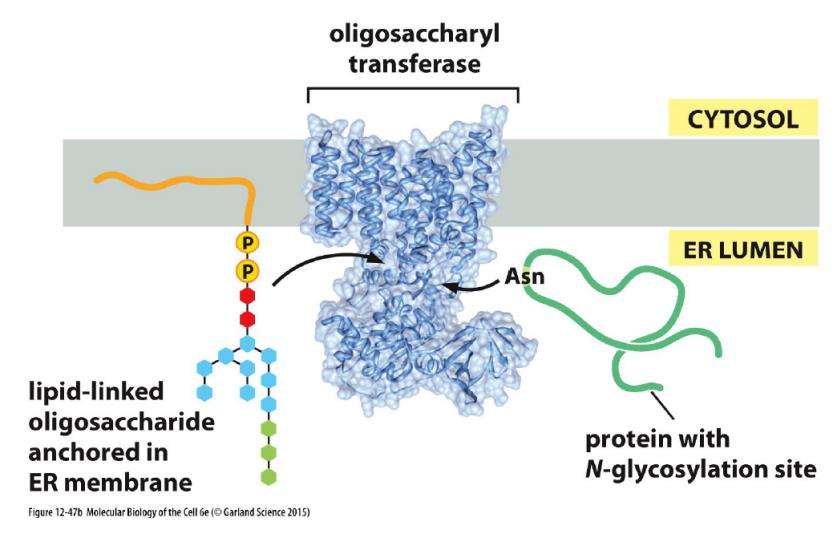
2. Oligosaccharide transfer catalyzed by oligosaccharyl transferase
It is catalyzed by oligosaccharyl transferase, whose active sites are exposed in the ER lumen only.
3. Modification of the oligosaccharide by removal of sugars
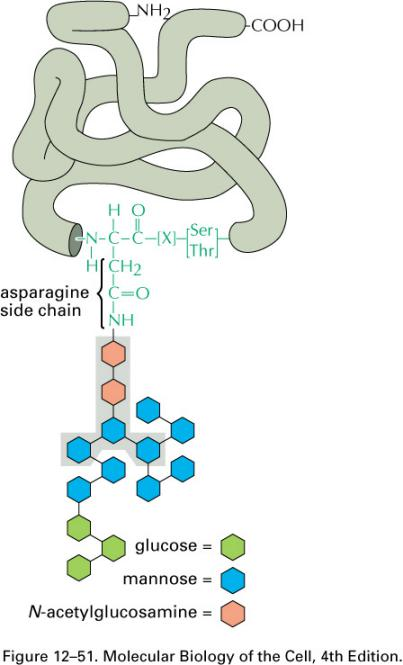
Finally modification of oligosaccharide
- Three glucoses and one mannose are removed sequentially in the ER.
4. Carbohydrate side chains sequentially modified by Golgi enzymes
Golgi enzymes modify N-linked glycosylation of proteins.
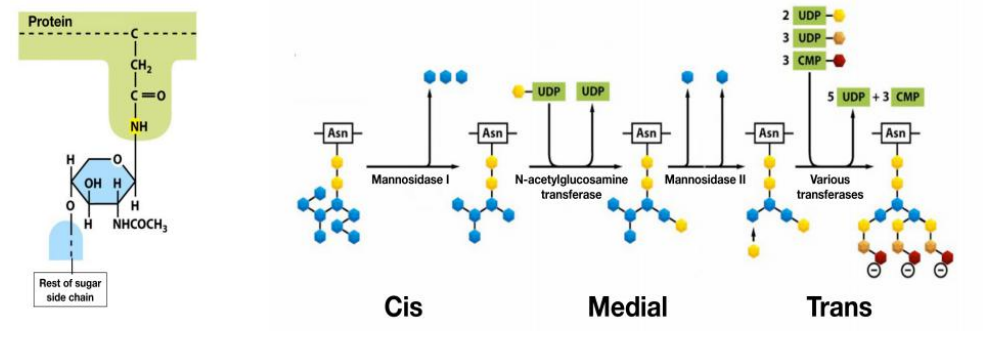
Golgi enzymes catalyze O-linked glycosylation of proteins
- O-linked glycosylation only happens in Golgi
Oligosaccharide processing in the ER and the Golgi
Summary: glycosylation
Glycosylation is the addition of sugars to proteins and occurs in the ER and Golgi.
- ER:
- Synthesis of the core portion of an N-linked oligosaccharide
- Golgi:
- Modification of N-linked oligosaccharides
- Synthesis of O-linked oligosaccharides
- Phosphorylation of mannose (on N-linked oligosaccharide) on proteins targeted for lysosomes (see lecture 11)
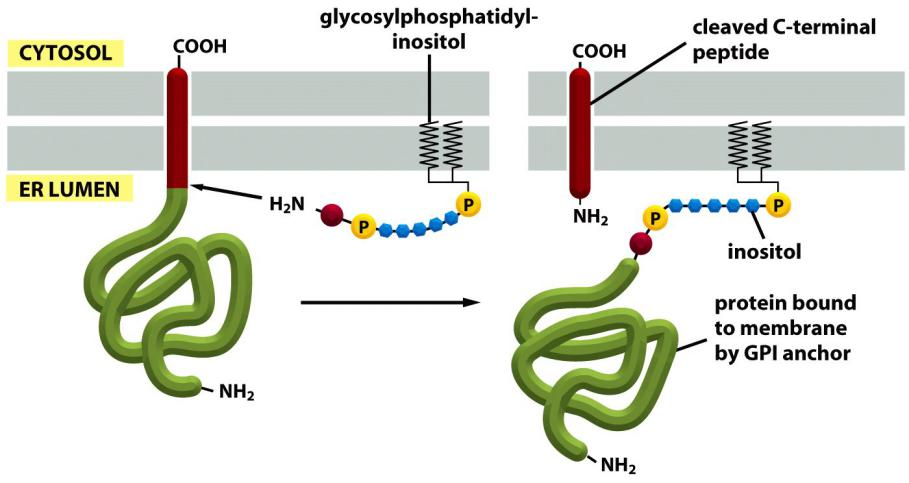
Attachment of the covalently linked GPI anchor
A related process is catalyzed by ER enzymes that covalently attach a glycosylphosphatidylinositol (GPI) anchor to the C-terminus of some membrane proteins destined for the plasma membrane.
This linkage forms in the lumen of the ER, where, at the same time, the transmembrane segment of the protein is cleaved off
A large number of plasma membrane proteins are modified in this way. Since they are attached to the exterior of the plasma membrane only by their GPI anchors
Sorting proteins in the Golgi
The Golgi apparatus receives lipids and proteins from the ER and dispatches them to various destinations, usually covalently modifying them en route.
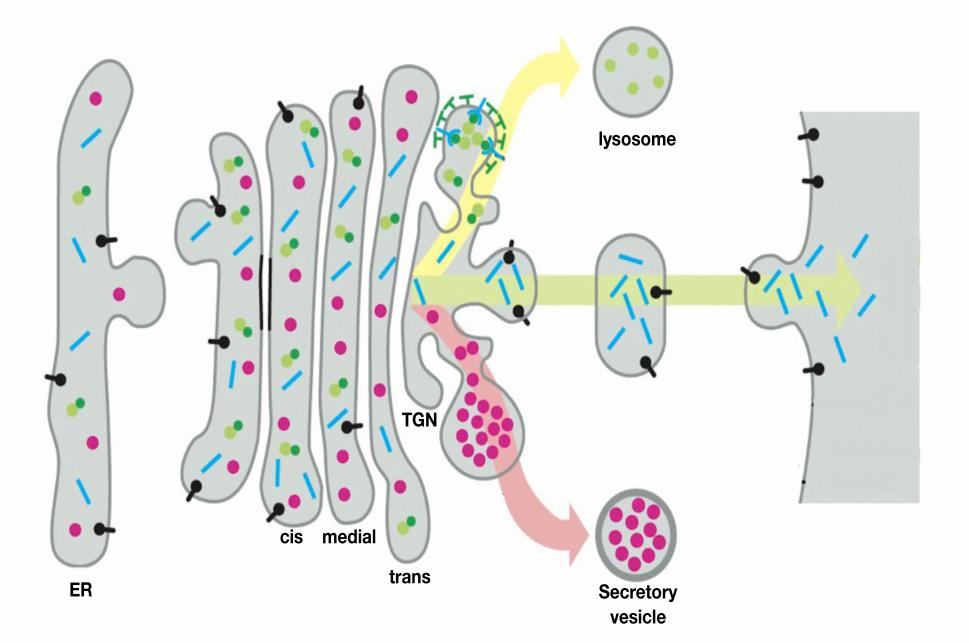
The ER is the main source for the lipids of the cell
Nearly all major lipids are synthesized in ER these include:
- Phospholipids
- Cholesterol
- Ceramide (precursor for Sphingomyelin)
Synthesis is on the cytosolic side of ER, where key enzymes are located.
Equal distribution for these lipids between both leaflets of ER after synthesis due to scramblase (混杂酶), which catalyzes rapid flip-flop.
1. The ER assembles most lipid bilayers
The ER membrane is the site of synthesis of nearly all of the cell’s major classes of lipids, including both phospholipids and cholesterol, required for the production of new cell membranes.
The major phospholipid made is phosphatidylcholine (PC), which can be formed in three steps from choline, two fatty acids, and glycerol phosphate
- phospholipid synthesis occurs exclusively in the cytosolic leaflet of the ER membrane
The synthesis of phosphatidylcholine (PC)
Similar mechanism for other major phospholipids
Equal distribution for these lipids between both leaflets of ER after synthesis due to scramblase, which catalyzes rapid flip-flop
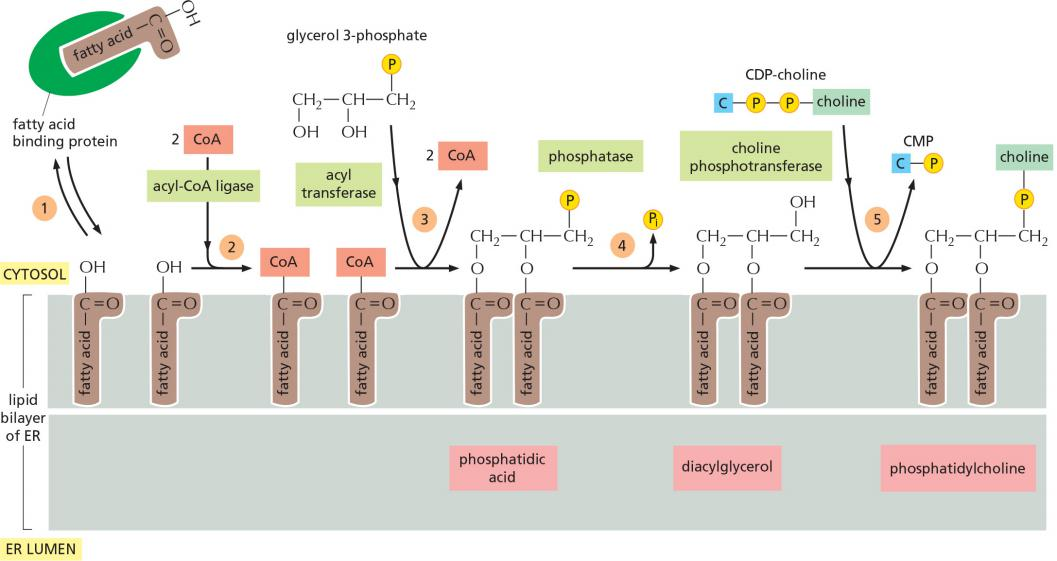
After arrival in the ER membrane and activation with CoA, acyl transferases successively add two fatty acids to glycerol phosphate (磷酸甘油)to produce phosphatidic acid(磷脂酸). Phosphatidic acid is sufficiently water-insoluble to remain in the lipid bilayer; it cannot be extracted from the bilayer by the fatty acid binding proteins
The later steps determine the head group of a newly formed lipid molecule and therefore the chemical nature of the bilayer, but they do not result in net membrane growth.
The two other major membrane phospholipids—phosphatidylethanolamine and phosphatidylserine (see Figure 10–3)—as well as the minor phospholipid phosphatidylinositol (PI), are all synthesized in this way.
2. Symmetry and asymmetry of lipids in ER and plasma membrane
In the ER, however, phospholipids equilibrate across the membrane within minutes, which is almost 100,000 times faster than can be accounted for by spontaneous“flip-flop.”
This rapid trans-bilayer movement is mediated by a poorly characterized phospholipid translocator called a scramblase, which non-selectively equilibrates phospholipids between the two leaflets of the lipid bilayer
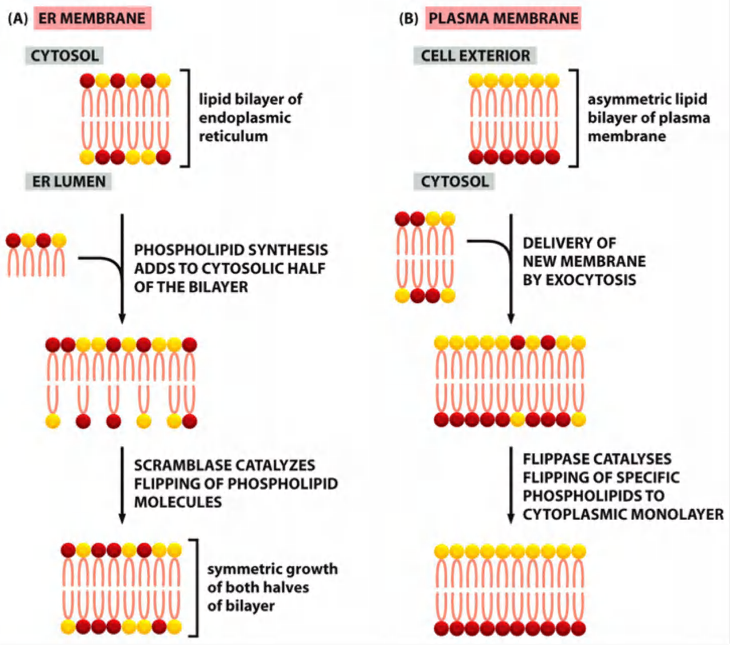
Phosphatidylserine (PS) and phosphatidylethanolamine (PE) are specifically “flipped” by an ATP-driven flippase (翻转酶)
The plasma membrane contains a different type of phospholipid translocator that belongs to the family of P-type pumps
These flippases specifically recognize those phospholipids that contain free amino groups in their head groups (phosphatidylserine and phosphatidylethanolamine) and transfers them from the extracellular to the cytosolic leaflet, using the energy of ATP hydrolysis
- The plasma membrane therefore has a highly asymmetric phospholipid composition, which is actively maintained by the flippases
- The resulting exposure of phosphatidylserine on the surface of apoptotic cells serves as a signal for phagocytic cells to ingest and degrade the dead cell.
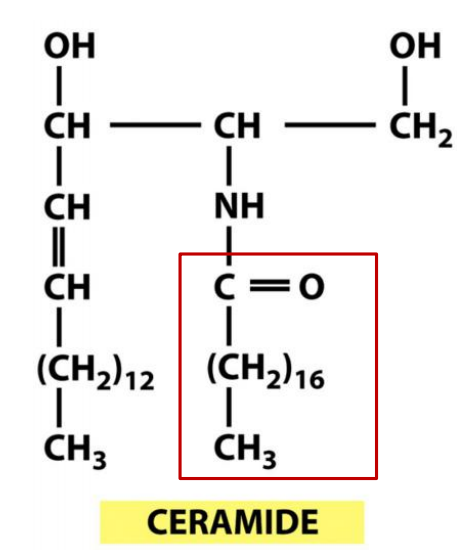
3. ER also synthesizes the sphingolipid ceramide
The ER also produces cholesterol and ceramide
- Ceramide is made by condensing the amino acid serine with a fatty acid to form the amino alcohol sphingosine, a second fatty acid is then covalently added to form ceramide.
- The ceramide is exported to the Golgi apparatus, where it serves as a precursor for the synthesis of two types of lipids: oligosaccharide chains are added to form glycosphingolipids, and phosphocholine head groups are transferred from phosphatidylcholine to other ceramide molecules to form sphingomyelin
Features
Ceramide is exported to the Golgi: precursor for the synthesis of
- glycosphingolipids
- sphingomyelin
Because they are produced by enzymes that have their active sites exposed to the Golgi lumen, they are found exclusively in the non-cytosolic leaflet of the lipid bilayers
4. Summary: ER functions
Functions of the Smooth ER
- Steroid hormone synthesis
- Detoxification in liver (lipid-soluble toxin)
- Release of glucose from liver
- Sequestering Ca2+ ions
Functions of the Rough ER
- Co-translational import of proteins destined for the ER-Golgi pathway
- Synthesis of membrane lipids and generation of lipid compositional asymmetry
- Protein glycosylation: Synthesis of the core portion of an N-linked oligosaccharide
The details of how lipid distribution between different membranes is catalyzed and regulated are not known. Water-soluble carrier proteins called phospholipid exchange proteins (or phospholipid transfer proteins) are thought to transfer individual phospholipid molecules between membranes, functioning much like fatty acid binding proteins that shepherd fatty acids through the cytosol